EpCAM-regulated intramembrane proteolysis induces a cancer stem cell-like gene signature in hepatitis B virus-infected hepatocytes
- PMID: 27238755
- PMCID: PMC5289705
- DOI: 10.1016/j.jhep.2016.05.022
EpCAM-regulated intramembrane proteolysis induces a cancer stem cell-like gene signature in hepatitis B virus-infected hepatocytes
Abstract
Background & aims: Hepatocytes in which the hepatitis B virus (HBV) is replicating exhibit loss of the chromatin modifying polycomb repressive complex 2 (PRC2), resulting in re-expression of specific, cellular PRC2-repressed genes. Epithelial cell adhesion molecule (EpCAM) is a PRC2-repressed gene, normally expressed in hepatic progenitors, but re-expressed in hepatic cancer stem cells (hCSCs). Herein, we investigated the functional significance of EpCAM re-expression in HBV-mediated hepatocarcinogenesis.
Methods: Employing molecular approaches (transfections, fluorescence-activated cell sorting, immunoblotting, qRT-PCR), we investigated the role of EpCAM-regulated intramembrane proteolysis (RIP) in HBV replicating cells in vitro, and in liver tumors from HBV X/c-myc mice and chronically HBV infected patients.
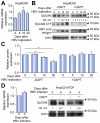
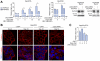
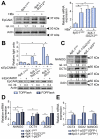
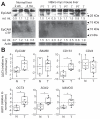
Results: EpCAM undergoes RIP in HBV replicating cells, activating canonical Wnt signaling. Transfection of Wnt-responsive plasmid expressing green fluorescent protein (GFP) identified a GFP + population of HBV replicating cells. These GFP+/Wnt+ cells exhibited cisplatin- and sorafenib-resistant growth resembling hCSCs, and increased expression of pluripotency genes NANOG, OCT4, SOX2, and hCSC markers BAMBI, CD44 and CD133. These genes are referred as EpCAM RIP and Wnt-induced hCSC-like gene signature. Interestingly, this gene signature is also overexpressed in liver tumors of X/c-myc bitransgenic mice. Clinically, a group of HBV-associated hepatocellular carcinomas was identified, exhibiting elevated expression of the hCSC-like gene signature and associated with reduced overall survival post-surgical resection.
Conclusions: The hCSC-like gene signature offers promise as prognostic tool for classifying subtypes of HBV-induced HCCs. Since EpCAM RIP and Wnt signaling drive expression of this hCSC-like signature, inhibition of these pathways can be explored as therapeutic strategy for this subtype of HBV-associated HCCs.
Lay summary: In this study, we provide evidence for a molecular mechanism by which chronic infection by the hepatitis B virus results in the development of poor prognosis liver cancer. Based on this mechanism our results suggest possible therapeutic interventions.
Keywords: BAMBI; CD133; CD44; EpCAM-regulated intramembrane proteolysis; Hepatitis B virus; Hepatocellular carcinoma; Pluripotency genes (NANOG, OCT4 and SOX2); SUZ12/polycomb repressive complex 2; Wnt signaling.
Copyright © 2016 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Figures



Comment in
-
Stemness of liver cancer: From hepatitis B virus to Wnt activation.J Hepatol. 2016 Nov;65(5):873-875. doi: 10.1016/j.jhep.2016.07.014. Epub 2016 Jul 19. J Hepatol. 2016. PMID: 27449917 No abstract available.
References
-
- Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22,707 men in Taiwan. Lancet. 1981;2:1129–1133. - PubMed
-
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. - PubMed
-
- Colombo M, Iavarone M. Role of antiviral treatment for HCC prevention. Best Pract Res Clin Gastroenterol. 2014;28:771–781. - PubMed
-
- Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608. e1591–e1592. - PubMed
-
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

