CRISPR-Mediated Drug-Target Validation Reveals Selective Pharmacological Inhibition of the RNA Helicase, eIF4A
- PMID: 27239032
- PMCID: PMC5315212
- DOI: 10.1016/j.celrep.2016.05.005
CRISPR-Mediated Drug-Target Validation Reveals Selective Pharmacological Inhibition of the RNA Helicase, eIF4A
Abstract
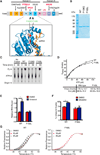
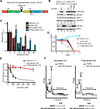
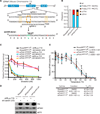
Targeting translation initiation is an emerging anti-neoplastic strategy that capitalizes on de-regulated upstream MAPK and PI3K-mTOR signaling pathways in cancers. A key regulator of translation that controls ribosome recruitment flux is eukaryotic initiation factor (eIF) 4F, a hetero-trimeric complex composed of the cap binding protein eIF4E, the scaffolding protein eIF4G, and the RNA helicase eIF4A. Small molecule inhibitors targeting eIF4F display promising anti-neoplastic activity in preclinical settings. Among these are some rocaglate family members that are well tolerated in vivo, deplete eIF4F of its eIF4A helicase subunit, have shown activity as single agents in several xenograft models, and can reverse acquired resistance to MAPK and PI3K-mTOR targeted therapies. Herein, we highlight the power of using genetic complementation approaches and CRISPR/Cas9-mediated editing for drug-target validation ex vivo and in vivo, linking the anti-tumor properties of rocaglates to eIF4A inhibition.
Keywords: CRISPR/Cas9; chemical biology; eIF4A; rocaglates; translational control.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

