Genetic engineering of AtAOX1a in Saccharomyces cerevisiae prevents oxidative damage and maintains redox homeostasis
- PMID: 27239435
- PMCID: PMC4821348
- DOI: 10.1002/2211-5463.12028
Genetic engineering of AtAOX1a in Saccharomyces cerevisiae prevents oxidative damage and maintains redox homeostasis
Abstract
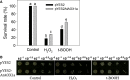
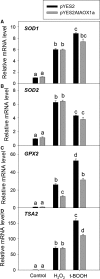
This study aimed to validate the physiological importance of Arabidopsis thaliana alternative oxidase 1a (AtAOX1a) in alleviating oxidative stress using Saccharomyces cerevisiae as a model organism. The AOX1a transformant (pYES2AtAOX1a) showed cyanide resistant and salicylhydroxamic acid (SHAM)-sensitive respiration, indicating functional expression of AtAOX1a in S. cerevisiae. After exposure to oxidative stress, pYES2AtAOX1a showed better survival and a decrease in reactive oxygen species (ROS) when compared to S. cerevisiae with empty vector (pYES2). Furthermore, pYES2AtAOX1a sustained growth by regulating GPX2 and/or TSA2, and cellular NAD (+)/NADH ratio. Thus, the expression of AtAOX1a in S. cerevisiae enhances its respiratory tolerance which, in turn, maintains cellular redox homeostasis and protects from oxidative damage.
Keywords: Saccharomyces cerevisiae; alternative oxidase 1a; oxidative stress; reactive oxygen species; redox homeostasis; respiration.
Figures



References
-
- McDonald AE, Vanlerberghe GC and Staples JF (2009) Alternative oxidase in animals: unique characteristics and taxonomic distribution. J Exp Biol 212, 2627–2634. - PubMed
-
- Moore AL and Siedow JN (1991) The regulation and nature of the cyanide‐resistant alternative oxidase of plant mitochondria. Biochim Biophys Acta 1059, 121–140. - PubMed
-
- Vanlerberghe GC and McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Biol 48, 703–734. - PubMed
-
- Millenaar F and Lambers H (2003) The alternative oxidase: in vivo regulation and function. Plant Biol 5, 2–15.
-
- Moore AL, Shiba T, Young L, Harada S, Kita K and Ito K (2013) Unraveling the heater: new insights into the structure of the alternative oxidase. Annu Rev Plant Biol 64, 637–663. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

