miRNA expression profiling of Epstein-Barr virus-associated NKTL cell lines by Illumina deep sequencing
- PMID: 27239439
- PMCID: PMC4821355
- DOI: 10.1002/2211-5463.12027
miRNA expression profiling of Epstein-Barr virus-associated NKTL cell lines by Illumina deep sequencing
Abstract
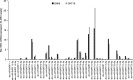
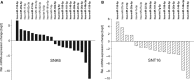
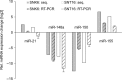
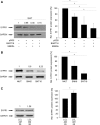
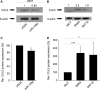
The aim of this work was to establish the microRNA profile of SNK6 and SNT16, two Epstein-Barr virus (EBV)-infected cell lines derived from nasal NK/T-cell lymphoma (NKTL). The oncogenic EBV is strongly associated with the pathogenesis of nasal and extranodal NK/T-cell lymphoma and expresses 44 mature microRNAs and two noncoding EBV-encoded RNAs (EBERs). miRNAs are 19-25nt noncoding RNAs that affect host and viral gene expression post-transcriptionally. Deregulated miRNA patterns are frequently linked to a variety of human cancers including lymphomas. miRNA profiling of the two NK/T cell lines vs. primary cells revealed 10 and 4 up-regulated and 10 and 12 down-regulated miRNAs in SNK6 and SNT16 cells respectively. The results were validated by qRT-PCR for selected miRNAs. Target gene analyses confirmed cullin 5 (CUL5) and sphingosin-1-phosphate receptor 1 (S1PR1) as targets for the down-regulated hsa-miR-148a and viral ebv-miR-BART16 respectively. As recently demonstrated for the regulation of IL1-alpha by miR-142-3p, coexpression of the EBERs selectively exerted corepression of S1PR1 by BART16 but not of CUL5 by miR-148a, indicating selective corepression by the EBERs.
Keywords: CUL5; Epstein‐Barr virus; S1PR1; ebv‐miR‐BART16; hsa‐miR‐148a; microRNA.
Figures



References
-
- Kieff E and Rickinson AE (2007) Epstein‐Barr Virus and its replication In Fields Virology. Vol. 2, 5th edn (Knipe D, Griffin DE, Lamb RA, Strauss SE, Howley PM, Martin MA. and Roizman B, eds), pp. 2603–2654. Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Rickinson AB and Kieff E (2007) Epstein‐Barr virus In Fields Virology. Vol. 2 (Knipe D, Griffin DE, Lamb RA, Strauss SE, Howley PM, Martin MA. and Roizman B, eds), pp. 2655–2700. Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Suzumiya J (2008) NK‐cell lymphoma: pathogenesis and clinical features. Rinsho Ketsueki 49, 545–552. - PubMed
-
- Jaffe ES, Chan JK, Su IJ, Frizzera G, Mori S, Feller AC and Ho FC (1996) Report of the Workshop on Nasal and Related Extranodal Angiocentric T/Natural Killer Cell Lymphomas. Definitions, differential diagnosis, and epidemiology. Am J Surg Pathol 20, 103–111. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

