Molecular profiling of dilated cardiomyopathy that progresses to heart failure
- PMID: 27239561
- PMCID: PMC4882118
- DOI: 10.1172/jci.insight.86898
Molecular profiling of dilated cardiomyopathy that progresses to heart failure
Abstract
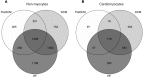
Dilated cardiomyopathy (DCM) is defined by progressive functional and structural changes. We performed RNA-seq at different stages of disease to define molecular signaling in the progression from pre-DCM hearts to DCM and overt heart failure (HF) using a genetic model of DCM (phospholamban missense mutation, PLNR9C/+). Pre-DCM hearts were phenotypically normal yet displayed proliferation of nonmyocytes (59% relative increase vs. WT, P = 8 × 10-4) and activation of proinflammatory signaling with notable cardiomyocyte-specific induction of a subset of profibrotic cytokines including TGFβ2 and TGFβ3. These changes progressed through DCM and HF, resulting in substantial fibrosis (17.6% of left ventricle [LV] vs. WT, P = 6 × 10-33). Cardiomyocytes displayed a marked shift in metabolic gene transcription: downregulation of aerobic respiration and subsequent upregulation of glucose utilization, changes coincident with attenuated expression of PPARα and PPARγ coactivators -1α (PGC1α) and -1β, and increased expression of the metabolic regulator T-box transcription factor 15 (Tbx15). Comparing DCM transcriptional profiles with those in hypertrophic cardiomyopathy (HCM) revealed similar and distinct molecular mechanisms. Our data suggest that cardiomyocyte-specific cytokine expression, early fibroblast activation, and the shift in metabolic gene expression are hallmarks of cardiomyopathy progression. Notably, key components of these profibrotic and metabolic networks were disease specific and distinguish DCM from HCM.
Figures


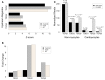



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

