Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection
- PMID: 27239795
- PMCID: PMC6522385
- DOI: 10.1038/nsmb.3236
Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection
Abstract
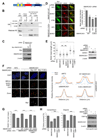
The opposing activities of 53BP1 and BRCA1 influence pathway choice in DNA double-strand-break repair. How BRCA1 counteracts the inhibitory effect of 53BP1 on DNA resection and homologous recombination is unknown. Here we identify the site of BRCA1-BARD1 required for priming ubiquitin transfer from E2∼ubiquitin and demonstrate that BRCA1-BARD1's ubiquitin ligase activity is required for repositioning 53BP1 on damaged chromatin. We confirm H2A ubiquitination by BRCA1-BARD1 and show that an H2A-ubiquitin fusion protein promotes DNA resection and repair in BARD1-deficient cells. BRCA1-BARD1's function in homologous recombination requires the chromatin remodeler SMARCAD1. SMARCAD1 binding to H2A-ubiquitin and optimal localization to sites of damage and activity in DNA repair requires its ubiquitin-binding CUE domains. SMARCAD1 is required for 53BP1 repositioning, and the need for SMARCAD1 in olaparib or camptothecin resistance is alleviated by 53BP1 loss. Thus, BRCA1-BARD1 ligase activity and subsequent SMARCAD1-dependent chromatin remodeling are critical regulators of DNA repair.
Figures




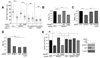


References
-
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

