A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses
- PMID: 27240195
- PMCID: PMC4942342
- DOI: 10.1038/nbt.3563
A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses
Abstract
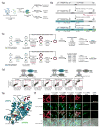
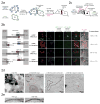
Intercellular protein-protein interactions (PPIs) enable communication between cells in diverse biological processes, including cell proliferation, immune responses, infection, and synaptic transmission, but they are challenging to visualize because existing techniques have insufficient sensitivity and/or specificity. Here we report a split horseradish peroxidase (sHRP) as a sensitive and specific tool for the detection of intercellular PPIs. The two sHRP fragments, engineered through screening of 17 cut sites in HRP followed by directed evolution, reconstitute into an active form when driven together by an intercellular PPI, producing bright fluorescence or contrast for electron microscopy. Fusing the sHRP fragments to the proteins neurexin (NRX) and neuroligin (NLG), which bind each other across the synaptic cleft, enabled sensitive visualization of synapses between specific sets of neurons, including two classes of synapses in the mouse visual system. sHRP should be widely applicable to studying mechanisms of communication between a variety of cell types.
Conflict of interest statement
Massachusetts Institute of Technology has filed a patent covering part of the information contained in this article.
Figures


References
-
- Feinberg EH, et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. - PubMed
-
- Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nature reviews Drug discovery. 2007;6:569–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

