An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy
- PMID: 27240197
- PMCID: PMC4942359
- DOI: 10.1038/nbt.3576
An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy
Abstract
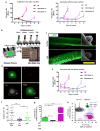
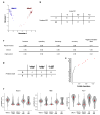
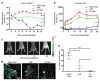
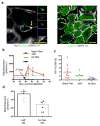
A promising therapeutic strategy for diverse genetic disorders involves transplantation of autologous stem cells that have been genetically corrected ex vivo. A major challenge in such approaches is a loss of stem cell potency during culture. Here we describe an artificial niche for maintaining muscle stem cells (MuSCs) in vitro in a potent, quiescent state. Using a machine learning method, we identified a molecular signature of quiescence and used it to screen for factors that could maintain mouse MuSC quiescence, thus defining a quiescence medium (QM). We also engineered muscle fibers that mimic the native myofiber of the MuSC niche. Mouse MuSCs maintained in QM on engineered fibers showed enhanced potential for engraftment, tissue regeneration and self-renewal after transplantation in mice. An artificial niche adapted to human cells similarly extended the quiescence of human MuSCs in vitro and enhanced their potency in vivo. Our approach for maintaining quiescence may be applicable to stem cells isolated from other tissues.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
Stem cells: Engineering an artificial niche for cell quiescence.Nat Rev Mol Cell Biol. 2016 Jun 22;17(7):398. doi: 10.1038/nrm.2016.82. Nat Rev Mol Cell Biol. 2016. PMID: 27329965 No abstract available.
References
-
- Montarras D, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

