DNA End Resection: Facts and Mechanisms
- PMID: 27240470
- PMCID: PMC4936662
- DOI: 10.1016/j.gpb.2016.05.002
DNA End Resection: Facts and Mechanisms
Abstract
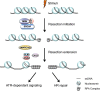
DNA double-strand breaks (DSBs), which arise following exposure to a number of endogenous and exogenous agents, can be repaired by either the homologous recombination (HR) or non-homologous end-joining (NHEJ) pathways in eukaryotic cells. A vital step in HR repair is DNA end resection, which generates a long 3' single-stranded DNA (ssDNA) tail that can invade the homologous DNA strand. The generation of 3' ssDNA is not only essential for HR repair, but also promotes activation of the ataxia telangiectasia and Rad3-related protein (ATR). Multiple factors, including the MRN/X complex, C-terminal-binding protein interacting protein (CtIP)/Sae2, exonuclease 1 (EXO1), Bloom syndrome protein (BLM)/Sgs1, DNA2 nuclease/helicase, and several chromatin remodelers, cooperate to complete the process of end resection. Here we review the basic machinery involved in DNA end resection in eukaryotic cells.
Keywords: Chromatin remodeling factors; DNA double-strand breaks; DNA end resection; Genome stability; Homologous recombination.
Copyright © 2016 The Authors. Production and hosting by Elsevier Ltd.. All rights reserved.
Figures
References
-
- van Gent D.C., Hoeijmakers J.H., Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. - PubMed
-
- Shiloh Y., Lehmann A.R. Maintaining integrity. Nat Cell Biol. 2004;6:923–928. - PubMed
-
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

