An Improved Method for High Quality Metagenomics DNA Extraction from Human and Environmental Samples
- PMID: 27240745
- PMCID: PMC4886217
- DOI: 10.1038/srep26775
An Improved Method for High Quality Metagenomics DNA Extraction from Human and Environmental Samples
Abstract
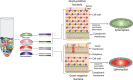
To explore the natural microbial community of any ecosystems by high-resolution molecular approaches including next generation sequencing, it is extremely important to develop a sensitive and reproducible DNA extraction method that facilitate isolation of microbial DNA of sufficient purity and quantity from culturable and uncultured microbial species living in that environment. Proper lysis of heterogeneous community microbial cells without damaging their genomes is a major challenge. In this study, we have developed an improved method for extraction of community DNA from different environmental and human origin samples. We introduced a combination of physical, chemical and mechanical lysis methods for proper lysis of microbial inhabitants. The community microbial DNA was precipitated by using salt and organic solvent. Both the quality and quantity of isolated DNA was compared with the existing methodologies and the supremacy of our method was confirmed. Maximum recovery of genomic DNA in the absence of substantial amount of impurities made the method convenient for nucleic acid extraction. The nucleic acids obtained using this method are suitable for different downstream applications. This improved method has been named as the THSTI method to depict the Institute where the method was developed.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

