Lessons from non-canonical splicing
- PMID: 27240813
- PMCID: PMC5154377
- DOI: 10.1038/nrg.2016.46
Lessons from non-canonical splicing
Abstract
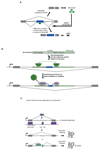
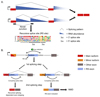
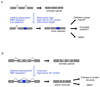
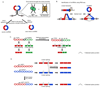
Recent improvements in experimental and computational techniques that are used to study the transcriptome have enabled an unprecedented view of RNA processing, revealing many previously unknown non-canonical splicing events. This includes cryptic events located far from the currently annotated exons and unconventional splicing mechanisms that have important roles in regulating gene expression. These non-canonical splicing events are a major source of newly emerging transcripts during evolution, especially when they involve sequences derived from transposable elements. They are therefore under precise regulation and quality control, which minimizes their potential to disrupt gene expression. We explain how non-canonical splicing can lead to aberrant transcripts that cause many diseases, and also how it can be exploited for new therapeutic strategies.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

