Analysis of the dynamic co-expression network of heart regeneration in the zebrafish
- PMID: 27241320
- PMCID: PMC4886216
- DOI: 10.1038/srep26822
Analysis of the dynamic co-expression network of heart regeneration in the zebrafish
Abstract
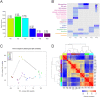
The zebrafish has the capacity to regenerate its heart after severe injury. While the function of a few genes during this process has been studied, we are far from fully understanding how genes interact to coordinate heart regeneration. To enable systematic insights into this phenomenon, we generated and integrated a dynamic co-expression network of heart regeneration in the zebrafish and linked systems-level properties to the underlying molecular events. Across multiple post-injury time points, the network displays topological attributes of biological relevance. We show that regeneration steps are mediated by modules of transcriptionally coordinated genes, and by genes acting as network hubs. We also established direct associations between hubs and validated drivers of heart regeneration with murine and human orthologs. The resulting models and interactive analysis tools are available at http://infused.vital-it.ch. Using a worked example, we demonstrate the usefulness of this unique open resource for hypothesis generation and in silico screening for genes involved in heart regeneration.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

