Transcription Profile of Aging and Cognition-Related Genes in the Medial Prefrontal Cortex
- PMID: 27242522
- PMCID: PMC4868850
- DOI: 10.3389/fnagi.2016.00113
Transcription Profile of Aging and Cognition-Related Genes in the Medial Prefrontal Cortex
Abstract
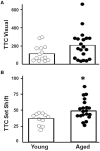
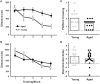
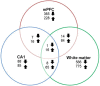
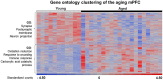
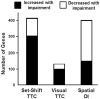
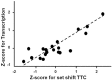
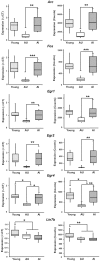
Cognitive function depends on transcription; however, there is little information linking altered gene expression to impaired prefrontal cortex function during aging. Young and aged F344 rats were characterized on attentional set shift and spatial memory tasks. Transcriptional differences associated with age and cognition were examined using RNA sequencing to construct transcriptomic profiles for the medial prefrontal cortex (mPFC), white matter, and region CA1 of the hippocampus. The results indicate regional differences in vulnerability to aging. Age-related gene expression in the mPFC was similar to, though less robust than, changes in the dorsolateral PFC of aging humans suggesting that aging processes may be similar. Importantly, the pattern of transcription associated with aging did not predict cognitive decline. Rather, increased mPFC expression of genes involved in regulation of transcription, including transcription factors that regulate the strength of excitatory and inhibitory inputs, and neural activity-related immediate-early genes was observed in aged animals that exhibit delayed set shift behavior. The specificity of impairment on a mPFC-dependent task, associated with a particular mPFC transcriptional profile indicates that impaired executive function involves altered transcriptional regulation and neural activity/plasticity processes that are distinct from that described for impaired hippocampal function.
Keywords: aging; cognitive flexibility; prefrontal cortex; set shifting task; transcription.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

