EP300 Protects from Light-Induced Retinopathy in Zebrafish
- PMID: 27242532
- PMCID: PMC4871856
- DOI: 10.3389/fphar.2016.00126
EP300 Protects from Light-Induced Retinopathy in Zebrafish
Abstract
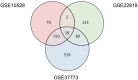
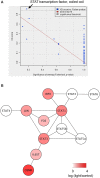
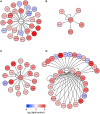
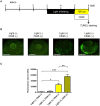
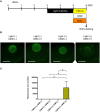
Exposure of rhodopsin to bright white light can induce photoreceptor cell damage and degeneration. However, a comprehensive understanding of the mechanisms underlying light-induced retinopathy remains elusive. In this study, we performed comparative transcriptome analysis of three rodent models of light-induced retinopathy, and we identified 37 genes that are dysregulated in all three models. Gene ontology analysis revealed that this gene set is significantly associated with a cytokine signaling axis composed of signal transducer and activator of transcription 1 and 3 (STAT1/3), interleukin 6 signal transducer (IL6ST), and oncostatin M receptor (OSMR). Furthermore, the analysis suggested that the histone acetyltransferase EP300 may be a key upstream regulator of the STAT1/3-IL6ST/OSMR axis. To examine the role of EP300 directly, we developed a larval zebrafish model of light-induced retinopathy. Using this model, we demonstrated that pharmacological inhibition of EP300 significantly increased retinal cell apoptosis, decreased photoreceptor cell outer segments, and increased proliferation of putative Müller cells upon exposure to intense light. These results suggest that EP300 may protect photoreceptor cells from light-induced damage and that activation of EP300 may be a novel therapeutic approach for the treatment of retinal degenerative diseases.
Keywords: EP300; STAT3; apoptosis; comparative transcriptome analysis; light-induced retinopathy; systems pharmacology; zebrafish.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

