Bovine Teat Microbiome Analysis Revealed Reduced Alpha Diversity and Significant Changes in Taxonomic Profiles in Quarters with a History of Mastitis
- PMID: 27242672
- PMCID: PMC4876361
- DOI: 10.3389/fmicb.2016.00480
Bovine Teat Microbiome Analysis Revealed Reduced Alpha Diversity and Significant Changes in Taxonomic Profiles in Quarters with a History of Mastitis
Abstract
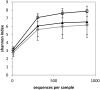
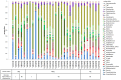
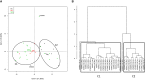
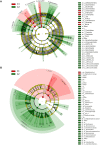
Mastitis is a mammary gland inflammatory disease often due to bacterial infections. Like many other infections, it used to be considered as a host-pathogen interaction driven by host and bacterial determinants. Until now, the involvement of the bovine mammary gland microbiota in the host-pathogen interaction has been poorly investigated, and mainly during the infectious episode. In this study, the bovine teat microbiome was investigated in 31 quarters corresponding to 27 animals, which were all free of inflammation at sampling time but which had different histories regarding mastitis: from no episode of mastitis on all the previous lactations (Healthy quarter, Hq) to one or several clinical mastitis events (Mastitic quarter, Mq). Several quarters whose status was unclear (possible history of subclinical mastitis) were classified as NDq. Total bacterial DNA was extracted from foremilk samples and swab samples of the teat canal. Taxonomic profiles were determined by pyrosequencing on 16s amplicons of the V3-4 region. Hq quarters showed a higher diversity compared to Mq ones (Shannon index: ~8 and 6, respectively). Clustering of the quarters based on their bacterial composition made it possible to separate Mq and Hq quarters into two separate clusters (C1 and C2, respectively). Discriminant analysis of taxonomic profiles between these clusters revealed several differences and allowed the identification of taxonomic markers in relation to mastitis history. C2 quarters were associated with a higher proportion of the Clostridia class (including genera such as Ruminococcus, Oscillospira, Roseburia, Dorea, etc.), the Bacteroidetes phylum (Prevotella, Bacteroides, Paludibacter, etc.), and the Bifidobacteriales order (Bifidobacterium), whereas C1 quarters showed a higher proportion of the Bacilli class (Staphylococcus) and Chlamydiia class. These results indicate that microbiota is altered in udders which have already developed mastitis, even far from the infectious episode. Microbiome alteration may have resulted from the infection itself and or the associated antibiotic treatment. Alternatively, differences in microbiome composition in udders with a history of mastitis may have occurred prior to the infection and even contributed to infection development. Further investigations on the dynamics of mammary gland microbiota will help to elucidate the contribution of this endogenous microbiota to the mammary gland health.
Keywords: bovine microbiome; bovine microbiota; dairy ruminant; dysbiosis; mammary gland; mastitis.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

