Selection and Validation of Housekeeping Genes as Reference for Gene Expression Studies in Pigeonpea (Cajanus cajan) under Heat and Salt Stress Conditions
- PMID: 27242803
- PMCID: PMC4865767
- DOI: 10.3389/fpls.2015.01071
Selection and Validation of Housekeeping Genes as Reference for Gene Expression Studies in Pigeonpea (Cajanus cajan) under Heat and Salt Stress Conditions
Abstract
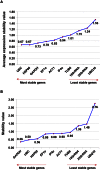
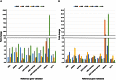
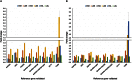
To identify stable housekeeping genes as a reference for expression analysis under heat and salt stress conditions in pigeonpea, the relative expression variation for 10 commonly used housekeeping genes (EF1α, UBQ10, GAPDH, 18Sr RNA, 25Sr RNA, TUB6, ACT1, IF4α, UBC, and HSP90) was studied in root, stem, and leaves tissues of Asha (ICPL 87119), a leading pigeonpea variety. Three statistical algorithms geNorm, NormFinder, and BestKeeper were used to define the stability of candidate genes. Under heat stress, UBC, HSP90, and GAPDH were found to be the most stable reference genes. In the case of salinity stress, GAPDH followed by UBC and HSP90 were identified to be the most stable reference genes. Subsequently, the above identified genes were validated using qRT-PCR based gene expression analysis of two universal stress-resposive genes namely uspA and uspB. The relative quantification of these two genes varied according to the internal controls (most stable, least stable, and combination of most stable and least stable housekeeping genes) and thus confirmed the choice as well as validation of internal controls in such experiments. The identified and validated housekeeping genes will facilitate gene expression studies under heat and salt stress conditions in pigeonpea.
Keywords: heat stress; housekeeping genes; quantitative real-time PCR; salt stress.
Figures


Similar articles
-
Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under drought stress conditions.PLoS One. 2015 Apr 7;10(4):e0122847. doi: 10.1371/journal.pone.0122847. eCollection 2015. PLoS One. 2015. PMID: 25849964 Free PMC article.
-
Gene expression analysis for selection and validation of suitable housekeeping gene(s) in cadmium exposed pigeonpea plants inoculated with arbuscular mycorrhizae.Plant Physiol Biochem. 2021 May;162:592-602. doi: 10.1016/j.plaphy.2021.03.024. Epub 2021 Mar 15. Plant Physiol Biochem. 2021. PMID: 33773234
-
Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculata.PeerJ. 2018 Jul 12;6:e5226. doi: 10.7717/peerj.5226. eCollection 2018. PeerJ. 2018. PMID: 30013853 Free PMC article.
-
Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses.Physiol Plant. 2015 Oct;155(2):138-148. doi: 10.1111/ppl.12302. Epub 2014 Nov 24. Physiol Plant. 2015. PMID: 25331743
-
New insights into the role of ribonuclease P protein subunit p30 from tumor to internal reference.Front Oncol. 2022 Oct 13;12:1018279. doi: 10.3389/fonc.2022.1018279. eCollection 2022. Front Oncol. 2022. PMID: 36313673 Free PMC article. Review.
Cited by
-
Assessment of changes in growth traits, oxidative stress parameters, and enzymatic and non-enzymatic antioxidant defense mechanisms in Lepidium draba plant under osmotic stress induced by polyethylene glycol.Protoplasma. 2020 Mar;257(2):459-473. doi: 10.1007/s00709-019-01457-0. Epub 2019 Nov 27. Protoplasma. 2020. PMID: 31776775
-
Comparative Transcriptomic Analysis on White and Blue Flowers of Platycodon grandiflorus to Elucidate Genes Involved in the Biosynthesis of Anthocyanins.Iran J Biotechnol. 2021 Jul 1;19(3):e2811. doi: 10.30498/ijb.2021.239899.2811. eCollection 2021 Jul. Iran J Biotechnol. 2021. PMID: 34825015 Free PMC article.
-
Drought and heat stress: insights into tolerance mechanisms and breeding strategies for pigeonpea improvement.Planta. 2024 Apr 15;259(5):123. doi: 10.1007/s00425-024-04401-6. Planta. 2024. PMID: 38622376 Review.
-
Selection of reliable reference genes for quantitative RT-PCR in garlic under salt stress.PeerJ. 2019 Jul 16;7:e7319. doi: 10.7717/peerj.7319. eCollection 2019. PeerJ. 2019. PMID: 31341748 Free PMC article.
-
Selection and validation of suitable reference genes for qRT-PCR analysis in pear leaf tissues under distinct training systems.PLoS One. 2018 Aug 23;13(8):e0202472. doi: 10.1371/journal.pone.0202472. eCollection 2018. PLoS One. 2018. PMID: 30138340 Free PMC article.
References
-
- Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of realtime quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 5245–5250. 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
-
- Chikelu M., Afza R., Jain S. M., Gregorio G. B., Zapata-Arias F. J. (2007). “Induced mutations for enhancing salinity tolerance in rice,” in Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops, eds Jenks M. A., Hasegawa P. M., Jain SM. (Dordrecht: Springer; ), 413–454.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

