Use of BABA and INA As Activators of a Primed State in the Common Bean (Phaseolus vulgaris L.)
- PMID: 27242854
- PMCID: PMC4870254
- DOI: 10.3389/fpls.2016.00653
Use of BABA and INA As Activators of a Primed State in the Common Bean (Phaseolus vulgaris L.)
Abstract
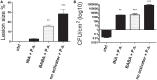
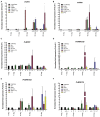
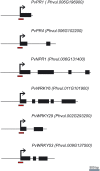
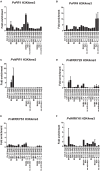
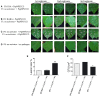
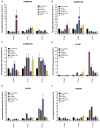
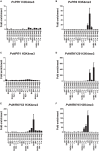
To survive in adverse conditions, plants have evolved complex mechanisms that "prime" their defense system to respond and adapt to stresses. Their competence to respond to such stresses fundamentally depends on its capacity to modulate the transcriptome rapidly and specifically. Thus, chromatin dynamics is a mechanism linked to transcriptional regulation and enhanced defense in plants. For example, in Arabidopsis, priming of the SA-dependent defense pathway is linked to histone lysine methylation. Such modifications could create a memory of the primary infection that is associated with an amplified gene response upon exposure to a second stress-stimulus. In addition, the priming status of a plant for induced resistance can be inherited to its offspring. However, analyses on the molecular mechanisms of generational and transgenerational priming in the common bean (Phaseolus vulagris L.), an economically important crop, are absent. Here, we provide evidence that resistance to P. syringae pv. phaseolicola infection was induced in the common bean with the synthetic priming activators BABA and INA. Resistance was assessed by evaluating symptom appearance, pathogen accumulation, changes in gene expression of defense genes, as well as changes in the H3K4me3 and H3K36me3 marks at the promoter-exon regions of defense-associated genes. We conclude that defense priming in the common bean occurred in response to BABA and INA and that these synthetic activators primed distinct genes for enhanced disease resistance. We hope that an understanding of the molecular changes leading to defense priming and pathogen resistance will provide valuable knowledge for producing disease-resistant crop varieties by exposing parental plants to priming activators, as well as to the development of novel plant protection chemicals that stimulate the plant's inherent disease resistance mechanisms.
Keywords: BABA; INA; common bean; epigenetics; priming.
Figures

Similar articles
-
Priming of seeds with INA and its transgenerational effect in common bean (Phaseolus vulgaris L.) plants.Plant Sci. 2021 Apr;305:110834. doi: 10.1016/j.plantsci.2021.110834. Epub 2021 Feb 5. Plant Sci. 2021. PMID: 33691968
-
Transgenerational Defense Priming for Crop Protection against Plant Pathogens: A Hypothesis.Front Plant Sci. 2017 May 4;8:696. doi: 10.3389/fpls.2017.00696. eCollection 2017. Front Plant Sci. 2017. PMID: 28523009 Free PMC article.
-
Immune Priming Triggers Cell Wall Remodeling and Increased Resistance to Halo Blight Disease in Common Bean.Plants (Basel). 2021 Jul 23;10(8):1514. doi: 10.3390/plants10081514. Plants (Basel). 2021. PMID: 34451558 Free PMC article.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Defense Priming: An Adaptive Part of Induced Resistance.Annu Rev Plant Biol. 2017 Apr 28;68:485-512. doi: 10.1146/annurev-arplant-042916-041132. Epub 2017 Feb 2. Annu Rev Plant Biol. 2017. PMID: 28226238 Review.
Cited by
-
ARABIDOPSIS HOMOLOG OF TRITHORAX1 impacts lateral root development by epigenetic regulation of targets involved in root system architecture.New Phytol. 2025 Sep;247(5):2180-2195. doi: 10.1111/nph.70349. Epub 2025 Jul 7. New Phytol. 2025. PMID: 40624794 Free PMC article.
-
Signalling mechanisms and agricultural applications of (Z)-3-hexenyl butyrate-mediated stomatal closure.Hortic Res. 2023 Nov 28;11(1):uhad248. doi: 10.1093/hr/uhad248. eCollection 2024 Jan. Hortic Res. 2023. PMID: 38239809 Free PMC article.
-
Nitric Oxide Plays an Important Role in β-Aminobutyric Acid-Induced Resistance to Botrytis cinerea in Tomato Plants.Plant Pathol J. 2020 Apr 1;36(2):121-132. doi: 10.5423/PPJ.OA.11.2019.0274. Plant Pathol J. 2020. PMID: 32296292 Free PMC article.
-
Transcriptional profiling of defense responses to Botrytis cinerea infection in leaves of Fragaria vesca plants soil-drenched with β-aminobutyric acid.Front Plant Sci. 2022 Dec 8;13:1025422. doi: 10.3389/fpls.2022.1025422. eCollection 2022. Front Plant Sci. 2022. PMID: 36570914 Free PMC article.
-
Epigenetic regulation of plant immunity: from chromatin codes to plant disease resistance.aBIOTECH. 2023 Mar 17;4(2):124-139. doi: 10.1007/s42994-023-00101-z. eCollection 2023 Jun. aBIOTECH. 2023. PMID: 37581024 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

