Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin
- PMID: 27242902
- PMCID: PMC4875998
- DOI: 10.1155/2016/8043983
Liposomal Systems as Nanocarriers for the Antiviral Agent Ivermectin
Abstract
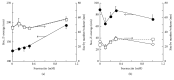
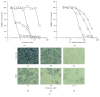
RNA virus infections can lead to the onset of severe diseases such as fever with haemorrhage, multiorgan failure, and mortality. The emergence and reemergence of RNA viruses continue to pose a significant public health threat worldwide with particular attention to the increasing incidence of flaviviruses, among others Dengue, West Nile Virus, and Yellow Fever viruses. Development of new and potent antivirals is thus urgently needed. Ivermectin, an already known antihelminthic drug, has shown potent effects in vitro on Flavivirus helicase, with EC50 values in the subnanomolar range for Yellow Fever and submicromolar EC50 for Dengue Fever, Japanese encephalitis, and tick-borne encephalitis viruses. However ivermectin is hampered in its application by pharmacokinetic problems (little solubility and high cytotoxicity). To overcome such problems we engineered different compositions of liposomes as ivermectin carriers characterizing and testing them on several cell lines for cytotoxicity. The engineered liposomes were less cytotoxic than ivermectin alone and they showed a significant increase of the antiviral activity in all the Dengue stains tested (1, 2, and S221). In the current study ivermectin is confirmed to be an effective potential antiviral and liposomes, as drug carriers, are shown to modulate the drug activity. All together the results represent a promising starting point for future improvement of ivermectin as antiviral and its delivery.
Figures










References
-
- Vasilakis N., Weaver S. C. The history and evolution of human dengue emergence. Advances in Virus Research. 2008;72:1–76. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

