Customized Steady-State Constraints for Parameter Estimation in Non-Linear Ordinary Differential Equation Models
- PMID: 27243005
- PMCID: PMC4863410
- DOI: 10.3389/fcell.2016.00041
Customized Steady-State Constraints for Parameter Estimation in Non-Linear Ordinary Differential Equation Models
Abstract
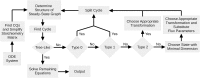
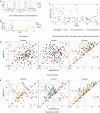
Ordinary differential equation models have become a wide-spread approach to analyze dynamical systems and understand underlying mechanisms. Model parameters are often unknown and have to be estimated from experimental data, e.g., by maximum-likelihood estimation. In particular, models of biological systems contain a large number of parameters. To reduce the dimensionality of the parameter space, steady-state information is incorporated in the parameter estimation process. For non-linear models, analytical steady-state calculation typically leads to higher-order polynomial equations for which no closed-form solutions can be obtained. This can be circumvented by solving the steady-state equations for kinetic parameters, which results in a linear equation system with comparatively simple solutions. At the same time multiplicity of steady-state solutions is avoided, which otherwise is problematic for optimization. When solved for kinetic parameters, however, steady-state constraints tend to become negative for particular model specifications, thus, generating new types of optimization problems. Here, we present an algorithm based on graph theory that derives non-negative, analytical steady-state expressions by stepwise removal of cyclic dependencies between dynamical variables. The algorithm avoids multiple steady-state solutions by construction. We show that our method is applicable to most common classes of biochemical reaction networks containing inhibition terms, mass-action and Hill-type kinetic equations. Comparing the performance of parameter estimation for different analytical and numerical methods of incorporating steady-state information, we show that our approach is especially well-tailored to guarantee a high success rate of optimization.
Keywords: biochemical reaction networks; multi-stability; multiplicity; non-linear ODE models; parameter estimation; positive solutions; steady-state; success rate.
Figures

Similar articles
-
Tailored parameter optimization methods for ordinary differential equation models with steady-state constraints.BMC Syst Biol. 2016 Aug 22;10(1):80. doi: 10.1186/s12918-016-0319-7. BMC Syst Biol. 2016. PMID: 27549154 Free PMC article.
-
A two-dimensional mathematical model of non-linear dual-sorption of percutaneous drug absorption.Biomed Eng Online. 2005 Jul 3;4:40. doi: 10.1186/1475-925X-4-40. Biomed Eng Online. 2005. PMID: 15992411 Free PMC article.
-
Efficient computation of adjoint sensitivities at steady-state in ODE models of biochemical reaction networks.PLoS Comput Biol. 2023 Jan 3;19(1):e1010783. doi: 10.1371/journal.pcbi.1010783. eCollection 2023 Jan. PLoS Comput Biol. 2023. PMID: 36595539 Free PMC article.
-
Classic and contemporary approaches to modeling biochemical reactions.Genes Dev. 2010 Sep 1;24(17):1861-75. doi: 10.1101/gad.1945410. Genes Dev. 2010. PMID: 20810646 Free PMC article. Review.
-
Scalable Inference of Ordinary Differential Equation Models of Biochemical Processes.Methods Mol Biol. 2019;1883:385-422. doi: 10.1007/978-1-4939-8882-2_16. Methods Mol Biol. 2019. PMID: 30547409 Review.
Cited by
-
Continuous analogue to iterative optimization for PDE-constrained inverse problems.Inverse Probl Sci Eng. 2018 Jul 10;27(6):710-734. doi: 10.1080/17415977.2018.1494167. eCollection 2019. Inverse Probl Sci Eng. 2018. PMID: 31057658 Free PMC article.
-
Parameterization of mechanistic models from qualitative data using an efficient optimal scaling approach.J Math Biol. 2020 Aug;81(2):603-623. doi: 10.1007/s00285-020-01522-w. Epub 2020 Jul 21. J Math Biol. 2020. PMID: 32696085 Free PMC article.
-
Basal MET phosphorylation is an indicator of hepatocyte dysregulation in liver disease.Mol Syst Biol. 2024 Mar;20(3):187-216. doi: 10.1038/s44320-023-00007-4. Epub 2024 Jan 12. Mol Syst Biol. 2024. PMID: 38216754 Free PMC article.
-
Tailored parameter optimization methods for ordinary differential equation models with steady-state constraints.BMC Syst Biol. 2016 Aug 22;10(1):80. doi: 10.1186/s12918-016-0319-7. BMC Syst Biol. 2016. PMID: 27549154 Free PMC article.
-
Benchmarking optimization methods for parameter estimation in large kinetic models.Bioinformatics. 2019 Mar 1;35(5):830-838. doi: 10.1093/bioinformatics/bty736. Bioinformatics. 2019. PMID: 30816929 Free PMC article.
References
-
- Feinberg M. (1979). Complex balancing in general kinetic systems. Arch. Rat. Mech. Anal. 49, 187–194. 10.1007/BF00255665 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

