The Statistical Value of Raw Fluorescence Signal in Luminex xMAP Based Multiplex Immunoassays
- PMID: 27243383
- PMCID: PMC4886638
- DOI: 10.1038/srep26996
The Statistical Value of Raw Fluorescence Signal in Luminex xMAP Based Multiplex Immunoassays
Abstract
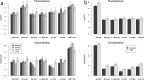
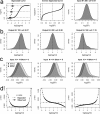
Tissue samples (plasma, saliva, serum or urine) from 169 patients classified as either normal or having one of seven possible diseases are analysed across three 96-well plates for the presences of 37 analytes using cytokine inflammation multiplexed immunoassay panels. Censoring for concentration data caused problems for analysis of the low abundant analytes. Using fluorescence analysis over concentration based analysis allowed analysis of these low abundant analytes. Mixed-effects analysis on the resulting fluorescence and concentration responses reveals a combination of censoring and mapping the fluorescence responses to concentration values, through a 5PL curve, changed observed analyte concentrations. Simulation verifies this, by showing a dependence on the mean florescence response and its distribution on the observed analyte concentration levels. Differences from normality, in the fluorescence responses, can lead to differences in concentration estimates and unreliable probabilities for treatment effects. It is seen that when fluorescence responses are normally distributed, probabilities of treatment effects for fluorescence based t-tests has greater statistical power than the same probabilities from concentration based t-tests. We add evidence that the fluorescence response, unlike concentration values, doesn't require censoring and we show with respect to differential analysis on the fluorescence responses that background correction is not required.
Figures


References
-
- Wong H. L. et al. Reproducibility and correlations of multiplex cytokine levels in asymptomatic persons. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 17, 3450–3456, 10.1158/1055-9965.EPI-08-0311 (2008). - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

