Single molecule analysis reveals reversible and irreversible steps during spliceosome activation
- PMID: 27244240
- PMCID: PMC4922858
- DOI: 10.7554/eLife.14166
Single molecule analysis reveals reversible and irreversible steps during spliceosome activation
Abstract
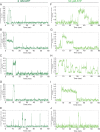
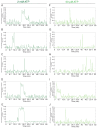
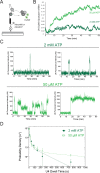
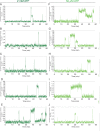
The spliceosome is a complex machine composed of small nuclear ribonucleoproteins (snRNPs) and accessory proteins that excises introns from pre-mRNAs. After assembly the spliceosome is activated for catalysis by rearrangement of subunits to form an active site. How this rearrangement is coordinated is not well-understood. During activation, U4 must be released to allow U6 conformational change, while Prp19 complex (NTC) recruitment is essential for stabilizing the active site. We used multi-wavelength colocalization single molecule spectroscopy to directly observe the key events in Saccharomyces cerevisiae spliceosome activation. Following binding of the U4/U6.U5 tri-snRNP, the spliceosome either reverses assembly by discarding tri-snRNP or proceeds to activation by irreversible U4 loss. The major pathway for NTC recruitment occurs after U4 release. ATP stimulates both the competing U4 release and tri-snRNP discard processes. The data reveal the activation mechanism and show that overall splicing efficiency may be maintained through repeated rounds of disassembly and tri-snRNP reassociation.
Keywords: RNA; S. cerevisiae; biochemistry; biophysics; fluorescence; single-molecule; snRNP; spliceosome; splicing; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



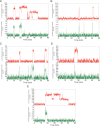
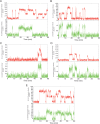
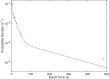

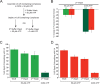


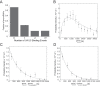
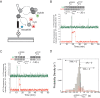


References
-
- Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, Guthrie C, Walter NG. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nature Structural & Molecular Biology. 2010;17:504–512. doi: 10.1038/nsmb.1767. - DOI - PMC - PubMed
-
- Boesler C, Rigo N, Agafonov DE, Kastner B, Urlaub H, Will CL, Lührmann R. Stable tri-snRNP integration is accompanied by a major structural rearrangement of the spliceosome that is dependent on Prp8 interaction with the 5' splice site. RNA. 2015;21:1993–2005. doi: 10.1261/rna.053991.115. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

