Modeling Effects of RNA on Capsid Assembly Pathways via Coarse-Grained Stochastic Simulation
- PMID: 27244559
- PMCID: PMC4887116
- DOI: 10.1371/journal.pone.0156547
Modeling Effects of RNA on Capsid Assembly Pathways via Coarse-Grained Stochastic Simulation
Abstract
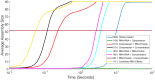
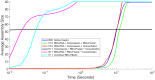
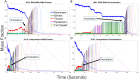
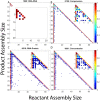
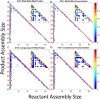
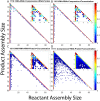
The environment of a living cell is vastly different from that of an in vitro reaction system, an issue that presents great challenges to the use of in vitro models, or computer simulations based on them, for understanding biochemistry in vivo. Virus capsids make an excellent model system for such questions because they typically have few distinct components, making them amenable to in vitro and modeling studies, yet their assembly can involve complex networks of possible reactions that cannot be resolved in detail by any current experimental technology. We previously fit kinetic simulation parameters to bulk in vitro assembly data to yield a close match between simulated and real data, and then used the simulations to study features of assembly that cannot be monitored experimentally. The present work seeks to project how assembly in these simulations fit to in vitro data would be altered by computationally adding features of the cellular environment to the system, specifically the presence of nucleic acid about which many capsids assemble. The major challenge of such work is computational: simulating fine-scale assembly pathways on the scale and in the parameter domains of real viruses is far too computationally costly to allow for explicit models of nucleic acid interaction. We bypass that limitation by applying analytical models of nucleic acid effects to adjust kinetic rate parameters learned from in vitro data to see how these adjustments, singly or in combination, might affect fine-scale assembly progress. The resulting simulations exhibit surprising behavioral complexity, with distinct effects often acting synergistically to drive efficient assembly and alter pathways relative to the in vitro model. The work demonstrates how computer simulations can help us understand how assembly might differ between the in vitro and in vivo environments and what features of the cellular environment account for these differences.
Conflict of interest statement
Figures




Similar articles
-
Modeling Viral Capsid Assembly: A Review of Computational Strategies and Applications.Cells. 2024 Dec 17;13(24):2088. doi: 10.3390/cells13242088. Cells. 2024. PMID: 39768179 Free PMC article. Review.
-
The Role of Packaging Sites in Efficient and Specific Virus Assembly.J Mol Biol. 2015 Jul 31;427(15):2451-2467. doi: 10.1016/j.jmb.2015.05.008. Epub 2015 May 16. J Mol Biol. 2015. PMID: 25986309 Free PMC article.
-
Role of electrostatics in the assembly pathway of a single-stranded RNA virus.J Virol. 2014 Sep;88(18):10472-9. doi: 10.1128/JVI.01044-14. Epub 2014 Jun 25. J Virol. 2014. PMID: 24965458 Free PMC article.
-
Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation.J Virol. 2016 May 27;90(12):5830-5844. doi: 10.1128/JVI.00394-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27076641 Free PMC article.
-
The interplay between capsid dynamics and pathogenesis in tripartite bromoviruses.Curr Opin Virol. 2021 Apr;47:45-51. doi: 10.1016/j.coviro.2020.12.005. Epub 2021 Jan 28. Curr Opin Virol. 2021. PMID: 33517133 Review.
Cited by
-
A Model for Viral Assembly around an Explicit RNA Sequence Generates an Implicit Fitness Landscape.Biophys J. 2017 Aug 8;113(3):506-516. doi: 10.1016/j.bpj.2017.06.037. Biophys J. 2017. PMID: 28793206 Free PMC article.
-
Quantitative computational models of molecular self-assembly in systems biology.Phys Biol. 2017 May 23;14(3):035003. doi: 10.1088/1478-3975/aa6cdc. Phys Biol. 2017. PMID: 28535149 Free PMC article. Review.
-
Coarse-Grained Models of RNA Nanotubes for Large Time Scale Studies in Biomedical Applications.Biomedicines. 2020 Jul 6;8(7):195. doi: 10.3390/biomedicines8070195. Biomedicines. 2020. PMID: 32640509 Free PMC article.
-
Modeling Viral Capsid Assembly: A Review of Computational Strategies and Applications.Cells. 2024 Dec 17;13(24):2088. doi: 10.3390/cells13242088. Cells. 2024. PMID: 39768179 Free PMC article. Review.
-
A method for efficient Bayesian optimization of self-assembly systems from scattering data.BMC Syst Biol. 2018 Jun 8;12(1):65. doi: 10.1186/s12918-018-0592-8. BMC Syst Biol. 2018. PMID: 29884203 Free PMC article.
References
-
- Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 1994:241: 59–67. - PubMed
-
- Zlotnick A, Johnson JM, Endres D. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry. 1999:38: 14644–14652. - PubMed
-
- Tsiang M, Niedziela-Majka A, Hung M, Jin D, Hu E, Yant S, et al. A trimer of dimers is the basic building block for human immunodeficiency virus-1 capsid assembly. Biochemistry. 2012:51: 4416–4428. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

