Combined physiological, transcriptome, and cis-regulatory element analyses indicate that key aspects of ripening, metabolism, and transcriptional program in grapes (Vitis vinifera L.) are differentially modulated accordingly to fruit size
- PMID: 27245662
- PMCID: PMC4886440
- DOI: 10.1186/s12864-016-2660-z
Combined physiological, transcriptome, and cis-regulatory element analyses indicate that key aspects of ripening, metabolism, and transcriptional program in grapes (Vitis vinifera L.) are differentially modulated accordingly to fruit size
Abstract
Background: In wine grape production, management practices have been adopted to optimize grape and wine quality attributes by producing, or screening for, berries of smaller size. Fruit size and composition are influenced by numerous factors that include both internal (e.g. berry hormone metabolism) and external (e.g. environment and cultural practices) factors. Combined physiological, biochemical, and transcriptome analyses were performed to improve our current understanding of metabolic and transcriptional pathways related to berry ripening and composition in berries of different sizes.
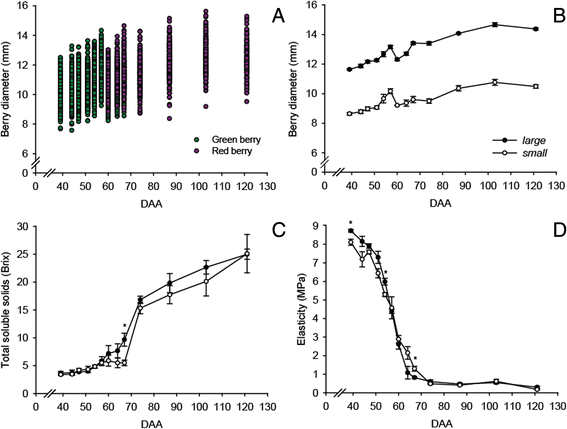
Results: The comparison of berry physiology between small and large berries throughout development (from 31 to 121 days after anthesis, DAA) revealed significant differences in firmness, the rate of softening, and sugar accumulation at specific developmental stages. Small berries had significantly higher skin to berry weight ratio, lower number of seeds per berry, and higher anthocyanin concentration compared to large berries. RNA-sequencing analyses of berry skins at 47, 74, 103, and 121 DAA revealed a total of 3482 differentially expressed genes between small and large berries. Abscisic acid, auxin, and ethylene hormone pathway genes were differentially modulated between berry sizes. Fatty acid degradation and stilbenoid pathway genes were upregulated at 47 DAA while cell wall degrading and modification genes were downregulated at 74 DAA in small compared to large berries. In the late ripening stage, concerted upregulation of the general phenylpropanoid and stilbenoid pathway genes and downregulation of flavonoid pathway genes were observed in skins of small compared to large berries. Cis-regulatory element analysis of differentially expressed hormone, fruit texture, flavor, and aroma genes revealed an enrichment of specific regulatory motifs related to bZIP, bHLH, AP2/ERF, NAC, MYB, and MADS-box transcription factors.
Conclusions: The study demonstrates that physiological and compositional differences between berries of different sizes parallel transcriptome changes that involve fruit texture, flavor, and aroma pathways. These results suggest that, in addition to direct effects brought about by differences in size, key aspects involved in the regulation of ripening likely contribute to different quality profiles between small and large berries.
Keywords: Aroma; Cell wall; Flavonoid; Grapevine; Hormone; Promoter; Quality; RNA-seq; Secondary metabolism; Transcriptomics.
Figures







References
-
- Singleton VL. Effects on red wine quality of removing juice before fermentation to simulate variation in berry size. Am J Enol Vitic. 1972;23:106–13.
-
- Matthews MA, Anderson MM. Fruit ripening in Vitis vinifera L.: responses to seasonal water deficits. Am J Enol Vitic. 1988;39:313–20.
-
- Coombe BG. Research on development and ripening of the grape berry. Am J Enol Vitic. 1992;43:101–10.
-
- Kuhn N, Guan L, Dai ZW, Wu BH, Lauvergeat V, Gomès E, Li SH, Godoy F, Arce-Johnson P, Delrot S. Berry ripening: recently heard through the grapevine. J Exp Bot. 2014;65:4543–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

