Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology
- PMID: 27245738
- PMCID: PMC4886425
- DOI: 10.1186/s12896-016-0277-6
Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology
Abstract
Background: The ATP-binding cassette (ABC) transporter gene superfamily is ubiquitous among extant organisms and prominently represented in plants. ABC transporters act to transport compounds across cellular membranes and are involved in a diverse range of biological processes. Thus, the applicability to biotechnology is vast, including cancer resistance in humans, drug resistance among vertebrates, and herbicide and other xenobiotic resistance in plants. In addition, plants appear to harbor the highest diversity of ABC transporter genes compared with any other group of organisms. This study applied transcriptome analysis to survey the kingdom-wide ABC transporter diversity in plants and suggest biotechnology applications of this diversity.
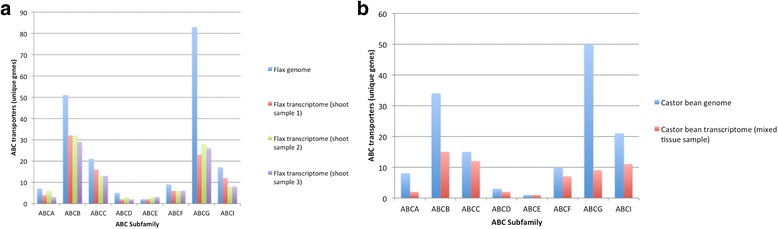
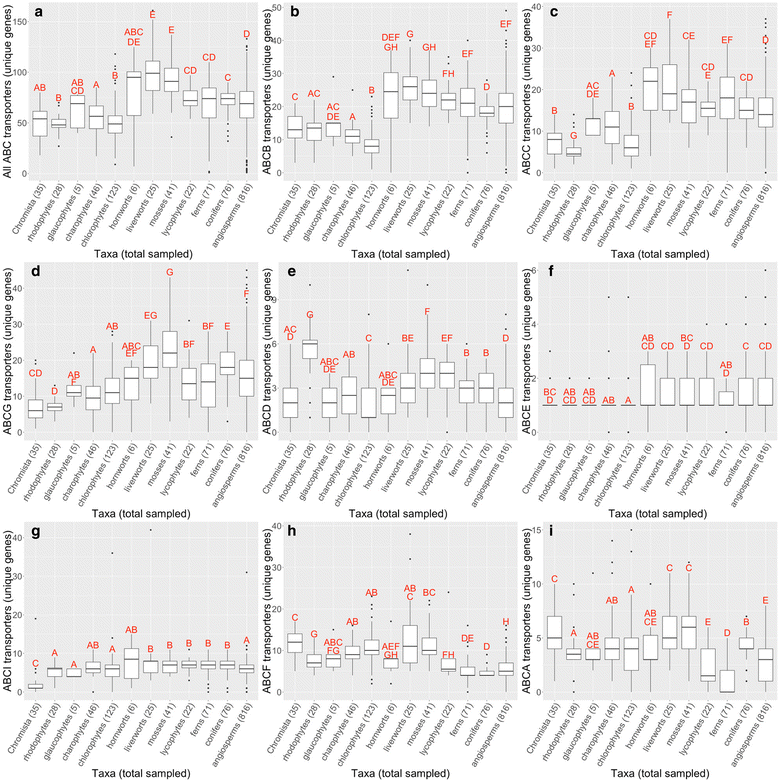
Results: We utilized sequence similarity-based informatics techniques to infer the identity of ABC transporter gene candidates from 1295 phylogenetically-diverse plant transcriptomes. A total of 97,149 putative (approximately 25 % were full-length) ABC transporter gene members were identified; each RNA-Seq library (plant sample) had 88 ± 30 gene members. As expected, simpler organisms, such as algae, had fewer unique members than vascular land plants. Differences were also noted in the richness of certain ABC transporter subfamilies. Land plants had more unique ABCB, ABCC, and ABCG transporter gene members on average (p < 0.005), and green algae, red algae, and bryophytes had significantly more ABCF transporter gene members (p < 0.005). Ferns had significantly fewer ABCA transporter gene members than all other plant groups (p < 0.005).
Conclusions: We present a transcriptomic overview of ABC transporter gene members across all major plant groups. An increase in the number of gene family members present in the ABCB, ABCC, and ABCD transporter subfamilies may indicate an expansion of the ABC transporter superfamily among green land plants, which include all crop species. The striking difference between the number of ABCA subfamily transporter gene members between ferns and other plant taxa is surprising and merits further investigation. Discussed is the potential exploitation of ABC transporters in plant biotechnology, with an emphasis on crops.
Keywords: ABC transporter; Computational biology; Taxonomic diversity; Transcriptomics.
Figures
Similar articles
-
Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp.PLoS One. 2019 Apr 30;14(4):e0215901. doi: 10.1371/journal.pone.0215901. eCollection 2019. PLoS One. 2019. PMID: 31039176 Free PMC article.
-
Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in sea lamprey and Japanese lamprey.BMC Genomics. 2015 Jun 6;16(1):436. doi: 10.1186/s12864-015-1677-z. BMC Genomics. 2015. PMID: 26047617 Free PMC article.
-
Genome-wide identification of ATP-binding cassette (ABC) transporters and conservation of their xenobiotic transporter function in the monogonont rotifer (Brachionus koreanus).Comp Biochem Physiol Part D Genomics Proteomics. 2017 Mar;21:17-26. doi: 10.1016/j.cbd.2016.10.003. Epub 2016 Oct 24. Comp Biochem Physiol Part D Genomics Proteomics. 2017. PMID: 27835832
-
Plant ABC transporters.Biochim Biophys Acta. 2000 May 1;1465(1-2):79-103. doi: 10.1016/s0005-2736(00)00132-2. Biochim Biophys Acta. 2000. PMID: 10748248 Review.
-
The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance.Insect Biochem Mol Biol. 2014 Feb;45:89-110. doi: 10.1016/j.ibmb.2013.11.001. Epub 2013 Nov 28. Insect Biochem Mol Biol. 2014. PMID: 24291285 Review.
Cited by
-
Compound heterozygous ABCA12 variants identified in a Chinese patient with congenital ichthyosiform erythroderma: Advancing genotype-phenotype correlations and literature review.Mol Genet Genomic Med. 2024 May;12(5):e2431. doi: 10.1002/mgg3.2431. Mol Genet Genomic Med. 2024. PMID: 38702946 Free PMC article. Review.
-
Identification and Characterization of ATP-Binding Cassette Transporters in Chlamydomonas reinhardtii.Mar Drugs. 2022 Sep 25;20(10):603. doi: 10.3390/md20100603. Mar Drugs. 2022. PMID: 36286426 Free PMC article.
-
Genome-Wide Identification of Barley ABC Genes and Their Expression in Response to Abiotic Stress Treatment.Plants (Basel). 2020 Sep 28;9(10):1281. doi: 10.3390/plants9101281. Plants (Basel). 2020. PMID: 32998428 Free PMC article.
-
Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp.PLoS One. 2019 Apr 30;14(4):e0215901. doi: 10.1371/journal.pone.0215901. eCollection 2019. PLoS One. 2019. PMID: 31039176 Free PMC article.
-
Drought and salinity induced changes in ecophysiology and proteomic profile of Parthenium hysterophorus.PLoS One. 2017 Sep 27;12(9):e0185118. doi: 10.1371/journal.pone.0185118. eCollection 2017. PLoS One. 2017. PMID: 28953916 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

