KCa1.1, a calcium-activated potassium channel subunit alpha 1, is targeted by miR-17-5p and modulates cell migration in malignant pleural mesothelioma
- PMID: 27245839
- PMCID: PMC4888473
- DOI: 10.1186/s12943-016-0529-z
KCa1.1, a calcium-activated potassium channel subunit alpha 1, is targeted by miR-17-5p and modulates cell migration in malignant pleural mesothelioma
Abstract
Background: Malignant pleural mesothelioma (MPM) is an aggressive, locally invasive, cancer elicited by asbestos exposure and almost invariably a fatal diagnosis. To date, we are one of the leading laboratory that compared microRNA expression profiles in MPM and normal mesothelium samples in order to identify dysregulated microRNAs with functional roles in mesothelioma. We interrogated a significant collection of MPM tumors and normal pleural samples in our biobank in search for novel therapeutic targets.
Methods: Utilizing mRNA-microRNA correlations based on differential gene expression using Gene Set Enrichment Analysis (GSEA), we systematically combined publicly available gene expression datasets with our own MPM data in order to identify candidate targets for MPM therapy.
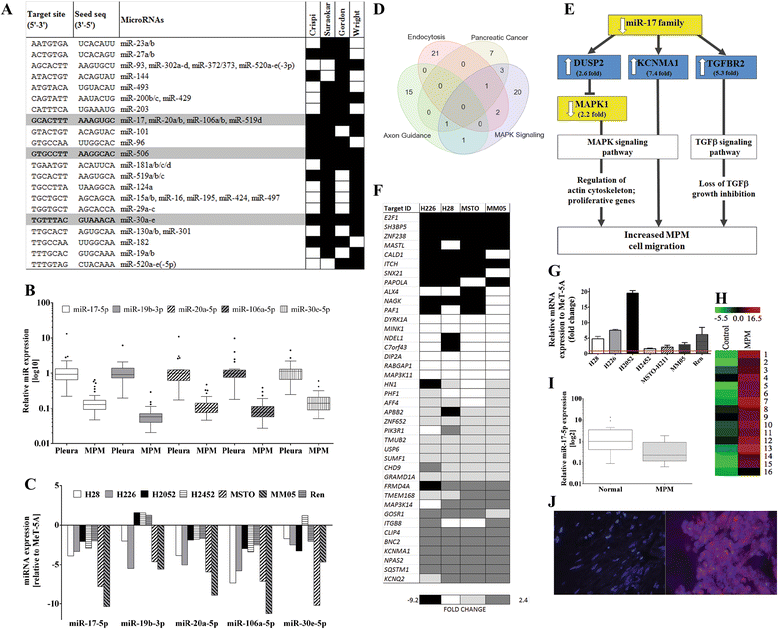
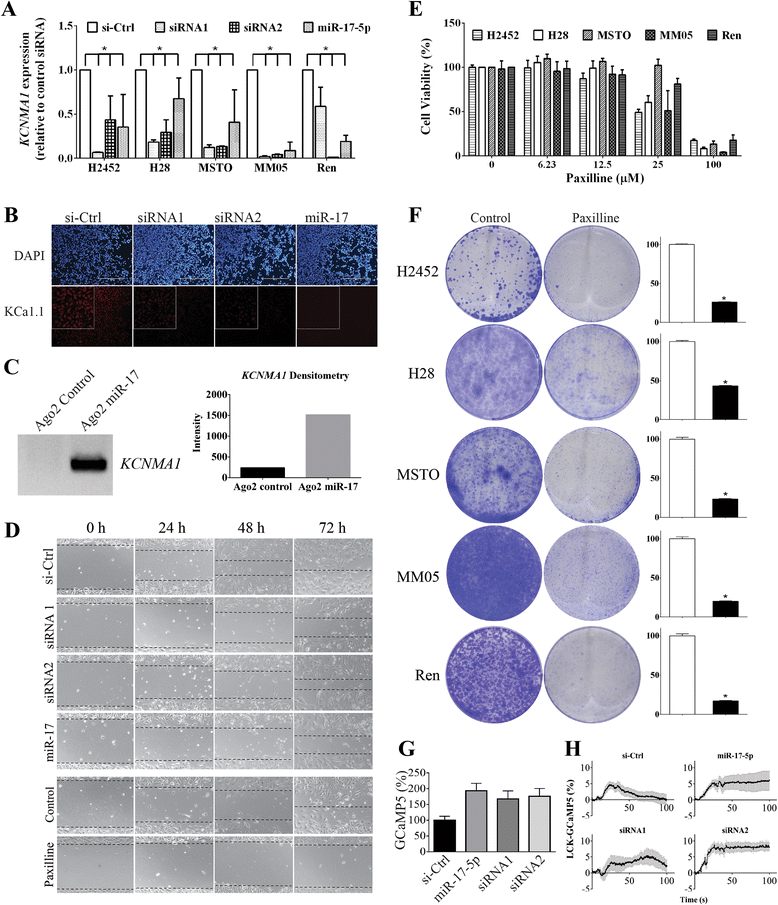
Results: We identified enrichment of target binding sites for the miR-17 and miR-30 families in both MPM tumors and cell lines. RT-qPCR revealed that members of both families were significantly downregulated in MPM tumors and cell lines. Interestingly, lower expression of miR-17-5p (P = 0.022) and miR-20a-5p (P = 0.026) was clearly associated with epithelioid histology. We interrogated the predicted targets of these differentially expressed microRNA families in MPM cell lines, and identified KCa1.1, a calcium-activated potassium channel subunit alpha 1 encoded by the KCNMA1 gene, as a target of miR-17-5p. KCa1.1 was overexpressed in MPM cells compared to the (normal) mesothelial line MeT-5A, and was also upregulated in patient tumor samples compared to normal mesothelium. Transfection of MPM cells with a miR-17-5p mimic or KCNMA1-specific siRNAs reduced mRNA expression of KCa1.1 and inhibited MPM cell migration. Similarly, treatment with paxilline, a small molecule inhibitor of KCa1.1, resulted in suppression of MPM cell migration.
Conclusion: These functional data implicating KCa1.1 in MPM cell migration support our integrative approach using MPM gene expression datasets to identify novel and potentially druggable targets.
Keywords: Integrative analysis; KCNMA1; KCa1.1; Mesothelioma; Therapeutic targets; miR-17-5p; microRNA.
Figures
Similar articles
-
Dysregulated Expression of the MicroRNA miR-137 and Its Target YBX1 Contribute to the Invasive Characteristics of Malignant Pleural Mesothelioma.J Thorac Oncol. 2018 Feb;13(2):258-272. doi: 10.1016/j.jtho.2017.10.016. Epub 2017 Nov 4. J Thorac Oncol. 2018. PMID: 29113949
-
Circulating microRNAs found dysregulated in ex-exposed asbestos workers and pleural mesothelioma patients as potential new biomarkers.Oncotarget. 2016 Dec 13;7(50):82700-82711. doi: 10.18632/oncotarget.12408. Oncotarget. 2016. PMID: 27716620 Free PMC article.
-
Protumorigenic effects of mir-145 loss in malignant pleural mesothelioma.Oncogene. 2014 Nov 13;33(46):5319-31. doi: 10.1038/onc.2013.476. Epub 2013 Nov 18. Oncogene. 2014. PMID: 24240684 Free PMC article.
-
Prognostic and Therapeutic Implications of MicroRNA in Malignant Pleural Mesothelioma.Microrna. 2016;5(1):12-18. doi: 10.2174/2211536605666160128151018. Microrna. 2016. PMID: 26817512 Review.
-
Secreted and Tissue miRNAs as Diagnosis Biomarkers of Malignant Pleural Mesothelioma.Int J Mol Sci. 2018 Feb 17;19(2):595. doi: 10.3390/ijms19020595. Int J Mol Sci. 2018. PMID: 29462963 Free PMC article. Review.
Cited by
-
Syntenin-1 is a promoter and prognostic marker of head and neck squamous cell carcinoma invasion and metastasis.Oncotarget. 2016 Dec 13;7(50):82634-82647. doi: 10.18632/oncotarget.13020. Oncotarget. 2016. PMID: 27811365 Free PMC article.
-
Transcriptional Repression and Protein Degradation of the Ca2+-Activated K+ Channel KCa1.1 by Androgen Receptor Inhibition in Human Breast Cancer Cells.Front Physiol. 2018 Apr 16;9:312. doi: 10.3389/fphys.2018.00312. eCollection 2018. Front Physiol. 2018. PMID: 29713287 Free PMC article.
-
Targeting microRNA to improve diagnostic and therapeutic approaches for malignant mesothelioma.Oncotarget. 2017 Aug 24;8(44):78193-78207. doi: 10.18632/oncotarget.20409. eCollection 2017 Sep 29. Oncotarget. 2017. PMID: 29100460 Free PMC article. Review.
-
Expression of potassium channel genes predicts clinical outcome in lung cancer.Korean J Physiol Pharmacol. 2019 Nov;23(6):529-537. doi: 10.4196/kjpp.2019.23.6.529. Epub 2019 Oct 24. Korean J Physiol Pharmacol. 2019. PMID: 31680775 Free PMC article.
-
3-Dimensional mesothelioma spheroids provide closer to natural pathophysiological tumor microenvironment for drug response studies.Front Oncol. 2022 Aug 26;12:973576. doi: 10.3389/fonc.2022.973576. eCollection 2022. Front Oncol. 2022. PMID: 36091141 Free PMC article.
References
-
- Scherpereel A, Astoul P, Baas P, Berghmans T, Clayson H, de Vuyst P, Dienemann H, Galateau-Salle F, Hennequin C, Hillerdal G, et al. Guidelines of the European Respiratory Society and the European Society of Thoracic Surgeons for the management of malignant pleural mesothelioma. Eur Respir J. 2010;35:479–495. doi: 10.1183/09031936.00063109. - DOI - PubMed
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. - DOI - PMC - PubMed
-
- Crispi S, Calogero RA, Santini M, Mellone P, Vincenzi B, Citro G, Vicidomini G, Fasano S, Meccariello R, Cobellis G, et al. Global gene expression profiling of human pleural mesotheliomas: identification of matrix metalloproteinase 14 (MMP-14) as potential tumour target. PLoS One. 2009;4:e7016. doi: 10.1371/journal.pone.0007016. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

