Genome stability versus transcript diversity
- PMID: 27246512
- PMCID: PMC4958514
- DOI: 10.1016/j.dnarep.2016.05.010
Genome stability versus transcript diversity
Abstract
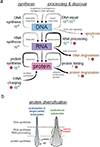
Our genome is protected from the introduction of mutations by high fidelity replication and an extensive network of DNA damage response and repair mechanisms. However, the expression of our genome, via RNA and protein synthesis, allows for more diversity in translating genetic information. In addition, the splicing process has become less stringent over evolutionary time allowing for a substantial increase in the diversity of transcripts generated. The result is a diverse transcriptome and proteome that harbor selective advantages over a more tightly regulated system. Here, we describe mechanisms in place that both safeguard the genome and promote translational diversity, with emphasis on post-transcriptional RNA processing.
Keywords: DNA damage response; DNA repair; Polymerase fidelity; Post-transcriptional RNA processing; Protein translation; Proteomic diversity; RNA splicing; RNA transcription.
Copyright © 2016 Elsevier B.V. All rights reserved.
Conflict of interest statement
None.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

