Functional Interaction between the Cytoplasmic ABC Protein LptB and the Inner Membrane LptC Protein, Components of the Lipopolysaccharide Transport Machinery in Escherichia coli
- PMID: 27246575
- PMCID: PMC4966433
- DOI: 10.1128/JB.00329-16
Functional Interaction between the Cytoplasmic ABC Protein LptB and the Inner Membrane LptC Protein, Components of the Lipopolysaccharide Transport Machinery in Escherichia coli
Abstract
The assembly of lipopolysaccharide (LPS) in the outer leaflet of the outer membrane (OM) requires the transenvelope Lpt (lipopolysaccharide transport) complex, made in Escherichia coli of seven essential proteins located in the inner membrane (IM) (LptBCFG), periplasm (LptA), and OM (LptDE). At the IM, LptBFG constitute an unusual ATP binding cassette (ABC) transporter, composed by the transmembrane LptFG proteins and the cytoplasmic LptB ATPase, which is thought to extract LPS from the IM and to provide the energy for its export across the periplasm to the cell surface. LptC is a small IM bitopic protein that binds to LptBFG and recruits LptA via its N- and C-terminal regions, and its role in LPS export is not completely understood. Here, we show that the expression level of lptB is a critical factor for suppressing lethality of deletions in the C-terminal region of LptC and the functioning of a hybrid Lpt machinery that carries Pa-LptC, the highly divergent LptC orthologue from Pseudomonas aeruginosa We found that LptB overexpression stabilizes C-terminally truncated LptC mutant proteins, thereby allowing the formation of a sufficient amount of stable IM complexes to support growth. Moreover, the LptB level seems also critical for the assembly of IM complexes carrying Pa-LptC which is otherwise defective in interactions with the E. coli LptFG components. Overall, our data suggest that LptB and LptC functionally interact and support a model whereby LptB plays a key role in the assembly of the Lpt machinery.
Importance: The asymmetric outer membrane (OM) of Gram-negative bacteria contains in its outer leaflet an unusual glycolipid, the lipopolysaccharide (LPS). LPS largely contributes to the peculiar permeability barrier properties of the OM that prevent the entry of many antibiotics, thus making Gram-negative pathogens difficult to treat. In Escherichia coli the LPS transporter (the Lpt machine) is made of seven essential proteins (LptABCDEFG) that form a transenvelope complex. Here, we show that increased expression of the membrane-associated ABC protein LptB can suppress defects of LptC, which participates in the formation of the periplasmic bridge. This reveals functional interactions between these two components and supports a role of LptB in the assembly of the Lpt machine.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

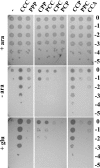
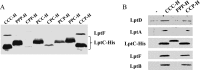

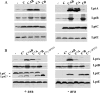


Similar articles
-
The Lack of the Essential LptC Protein in the Trans-Envelope Lipopolysaccharide Transport Machine Is Circumvented by Suppressor Mutations in LptF, an Inner Membrane Component of the Escherichia coli Transporter.PLoS One. 2016 Aug 16;11(8):e0161354. doi: 10.1371/journal.pone.0161354. eCollection 2016. PLoS One. 2016. PMID: 27529623 Free PMC article.
-
The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains.J Bacteriol. 2013 Mar;195(5):1100-8. doi: 10.1128/JB.02057-12. Epub 2013 Jan 4. J Bacteriol. 2013. PMID: 23292770 Free PMC article.
-
New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: LptA-LptC interaction and LptA stability as sensors of a properly assembled transenvelope complex.J Bacteriol. 2011 Mar;193(5):1042-53. doi: 10.1128/JB.01037-10. Epub 2010 Dec 17. J Bacteriol. 2011. PMID: 21169485 Free PMC article.
-
Structural insight into lipopolysaccharide transport from the Gram-negative bacterial inner membrane to the outer membrane.Biochim Biophys Acta Mol Cell Biol Lipids. 2017 Nov;1862(11):1461-1467. doi: 10.1016/j.bbalip.2017.08.003. Epub 2017 Aug 15. Biochim Biophys Acta Mol Cell Biol Lipids. 2017. PMID: 28821406 Review.
-
Structural Basis for the Lipopolysaccharide Export Activity of the Bacterial Lipopolysaccharide Transport System.Int J Mol Sci. 2018 Sep 10;19(9):2680. doi: 10.3390/ijms19092680. Int J Mol Sci. 2018. PMID: 30201863 Free PMC article. Review.
Cited by
-
The Lack of the Essential LptC Protein in the Trans-Envelope Lipopolysaccharide Transport Machine Is Circumvented by Suppressor Mutations in LptF, an Inner Membrane Component of the Escherichia coli Transporter.PLoS One. 2016 Aug 16;11(8):e0161354. doi: 10.1371/journal.pone.0161354. eCollection 2016. PLoS One. 2016. PMID: 27529623 Free PMC article.
-
The lipopolysaccharide transport (Lpt) machinery: A nonconventional transporter for lipopolysaccharide assembly at the outer membrane of Gram-negative bacteria.J Biol Chem. 2017 Nov 3;292(44):17981-17990. doi: 10.1074/jbc.R117.802512. Epub 2017 Sep 6. J Biol Chem. 2017. PMID: 28878019 Free PMC article. Review.
-
Characterization of and lipopolysaccharide binding to the E. coli LptC protein dimer.Protein Sci. 2018 Feb;27(2):381-389. doi: 10.1002/pro.3322. Epub 2017 Oct 28. Protein Sci. 2018. PMID: 29024084 Free PMC article.
-
More than Rotating Flagella: Lipopolysaccharide as a Secondary Receptor for Flagellotropic Phage 7-7-1.J Bacteriol. 2018 Sep 10;200(19):e00363-18. doi: 10.1128/JB.00363-18. Print 2018 Oct 1. J Bacteriol. 2018. PMID: 30012730 Free PMC article.
-
Phillygenin Inhibits Helicobacter pylori by Preventing Biofilm Formation and Inducing ATP Leakage.Front Microbiol. 2022 Apr 28;13:863624. doi: 10.3389/fmicb.2022.863624. eCollection 2022. Front Microbiol. 2022. PMID: 35572695 Free PMC article.
References
-
- Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Dehò G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

