Circular RNAs: analysis, expression and potential functions
- PMID: 27246710
- PMCID: PMC4920157
- DOI: 10.1242/dev.128074
Circular RNAs: analysis, expression and potential functions
Abstract
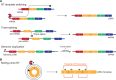
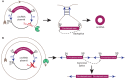
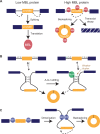
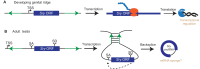
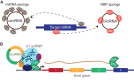
Just a few years ago, it had been assumed that the dominant RNA isoforms produced from eukaryotic genes were variants of messenger RNA, functioning as intermediates in gene expression. In early 2012, however, a surprising discovery was made: circular RNA (circRNA) was shown to be a transcriptional product in thousands of human and mouse genes and in hundreds of cases constituted the dominant RNA isoform. Subsequent studies revealed that the expression of circRNAs is developmentally regulated, tissue and cell-type specific, and shared across the eukaryotic tree of life. These features suggest important functions for these molecules. Here, we describe major advances in the field of circRNA biology, focusing on the regulation of and functional roles played by these molecules.
Keywords: CircRNA; Development; Non-coding RNA; Regulation; Splicing.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


References
-
- Artero R., Prokop A., Paricio N., Begemann G., Pueyo I., Mlodzik M., Perez-Alonso M. and Baylies M. K. (1998). The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev. Biol. 195, 131-143. 10.1006/dbio.1997.8833 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

