The C-Terminal RGG Domain of Human Lsm4 Promotes Processing Body Formation Stimulated by Arginine Dimethylation
- PMID: 27247266
- PMCID: PMC4985932
- DOI: 10.1128/MCB.01102-15
The C-Terminal RGG Domain of Human Lsm4 Promotes Processing Body Formation Stimulated by Arginine Dimethylation
Abstract
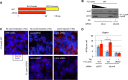
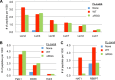
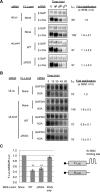
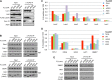
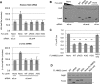
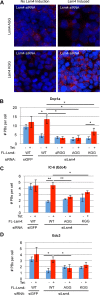
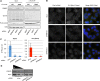
Processing bodies (PBs) are conserved cytoplasmic aggregations of translationally repressed mRNAs assembled with mRNA decay factors. The aggregation of mRNA-protein (mRNP) complexes into PBs involves interactions between low-complexity regions of protein components of the mRNPs. In Saccharomyces cerevisiae, the carboxy (C)-terminal Q/N-rich domain of the Lsm4 subunit of the Lsm1-7 complex plays an important role in PB formation, but the C-terminal domain of Lsm4 in most eukaryotes is an RGG domain rather than Q/N rich. Here we show that the Lsm4 RGG domain promotes PB accumulation in human cells and that symmetric dimethylation of arginines within the RGG domain stimulates this process. A mutant Lsm4 protein lacking the RGG domain failed to rescue PB formation in cells depleted of endogenous Lsm4, although this mutant protein retained the ability to assemble with Lsm1-7, associate with decapping factors, and promote mRNA decay and translational repression. Mutation of the symmetrically dimethylated arginines within the RGG domain impaired the ability of Lsm4 to promote PB accumulation. Depletion of PRMT5, the primary protein arginine methyltransferase responsible for symmetric arginine dimethylation, including Lsm4, resulted in loss of PBs. We also uncovered the histone acetyltransferase 1 (HAT1)-RBBP7 lysine acetylase complex as an interaction partner of the Lsm4 RGG domain but found no evidence of a role for this complex in PB metabolism. Together, our findings suggest a stimulatory role for posttranslational modifications in PB accumulation and raise the possibility that mRNP dynamics are posttranslationally regulated.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
