Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins
- PMID: 27247276
- PMCID: PMC4889330
- DOI: 10.7554/eLife.13325
Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins
Abstract
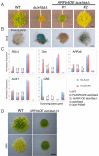
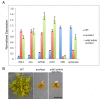
The coordinated action of the auxin-sensitive Aux/IAA transcriptional repressors and ARF transcription factors produces complex gene-regulatory networks in plants. Despite their importance, our knowledge of these two protein families is largely based on analysis of stabilized forms of the Aux/IAAs, and studies of a subgroup of ARFs that function as transcriptional activators. To understand how auxin regulates gene expression we generated a Physcomitrella patens line that completely lacks Aux/IAAs. Loss of the repressors causes massive changes in transcription with misregulation of over a third of the annotated genes. Further, we find that the aux/iaa mutant is blind to auxin indicating that auxin regulation of transcription occurs exclusively through Aux/IAA function. We used the aux/iaa mutant as a simplified platform for studies of ARF function and demonstrate that repressing ARFs regulate auxin-induced genes and fine-tune their expression. Further the repressing ARFs coordinate gene induction jointly with activating ARFs and the Aux/IAAs.
Keywords: ARFs; Aux/IAAs; Physcomitrella patens; auxin; chromosomes; genes; plant biology; plant development.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures













References
-
- Bennett TA, Liu MM, Aoyama T, Bierfreund NM, Braun M, Coudert Y, Dennis RJ, O'Connor D, Wang XY, White CD, Decker EL, Reski R, Harrison CJ. Plasma membrane-targeted PIN proteins drive shoot development in a moss. Current Biology. 2014;24:2776–2785. doi: 10.1016/j.cub.2014.09.054. - DOI - PMC - PubMed

