Whole-cell conversion of l-glutamic acid into gamma-aminobutyric acid by metabolically engineered Escherichia coli
- PMID: 27247887
- PMCID: PMC4864792
- DOI: 10.1186/s40064-016-2217-2
Whole-cell conversion of l-glutamic acid into gamma-aminobutyric acid by metabolically engineered Escherichia coli
Abstract
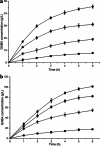
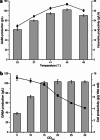
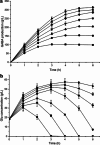
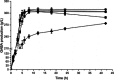
A simple and high efficient way for the synthesis of gamma-aminobutyric acid (GABA) was developed by using engineered Escherichia coli as whole-cell biocatalyst from l-glutamic acid (l-Glu). Codon optimization of Lactococcus lactis GadB showed the best performance on GABA production when middle copy-number plasmid was used as expression vector in E. coli BW25113. The highest production of GABA reached 308.96 g L(-1) with 99.9 mol% conversion within 12 h, when E. coli ΔgabAB (pRB-lgadB) concentrated to an OD600 of 15 in 3 M l-Glu at 45 °C. Furthermore, the strain could be reused at least three cycles in 2 M crude l-Glu with an average productivity of 40.94 g L(-1) h(-1). The total GABA yield reached 614.15 g L(-1) with a molar yield over 99 %, which represented the highest GABA production ever reported. The whole-cell bioconversion system allowed us to achieve a promising cost-effective resource for GABA in industrial application.
Keywords: Bioconversion; Escherichia coli; Gamma-aminobutyric acid; Glutamate decarboxylase; Whole-cell biocatalyst.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

