Intragenic Locus in Human PIWIL2 Gene Shares Promoter and Enhancer Functions
- PMID: 27248499
- PMCID: PMC4889060
- DOI: 10.1371/journal.pone.0156454
Intragenic Locus in Human PIWIL2 Gene Shares Promoter and Enhancer Functions
Abstract
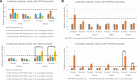
Recently, more evidence supporting common nature of promoters and enhancers has been accumulated. In this work, we present data on chromatin modifications and non-polyadenylated transcription characteristic for enhancers as well as results of in vitro luciferase reporter assays suggesting that PIWIL2 alternative promoter in exon 7 also functions as an enhancer for gene PHYHIP located 60Kb upstream. This finding of an intragenic enhancer serving as a promoter for a shorter protein isoform implies broader impact on understanding enhancer-promoter networks in regulation of gene expression.
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

