Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures
- PMID: 27249102
- PMCID: PMC4931852
- DOI: 10.18632/aging.100972
Epigenetic age predictions based on buccal swabs are more precise in combination with cell type-specific DNA methylation signatures
Abstract
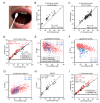
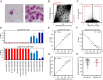
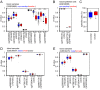
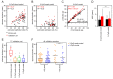
Aging is reflected by highly reproducible DNA methylation (DNAm) changes that open new perspectives for estimation of chronological age in legal medicine. DNA can be harvested non-invasively from cells at the inside of a person's cheek using buccal swabs - but these specimens resemble heterogeneous mixtures of buccal epithelial cells and leukocytes with different epigenetic makeup. In this study, we have trained an age predictor based on three age-associated CpG sites (associated with the genesPDE4C, ASPA, and ITGA2B) for swab samples to reach a mean absolute deviation (MAD) between predicted and chronological age of 4.3 years in a training set and of 7.03 years in a validation set. Subsequently, the composition of buccal epithelial cells versus leukocytes was estimated by two additional CpGs (associated with the genes CD6 and SERPINB5). Results of this "Buccal-Cell-Signature" correlated with cell counts in cytological stains (R2 = 0.94). Combination of cell type-specific and age-associated CpGs into one multivariate model enabled age predictions with MADs of 5.09 years and 5.12 years in two independent validation sets. Our results demonstrate that the cellular composition in buccal swab samples can be determined by DNAm at two cell type-specific CpGs to improve epigenetic age predictions.
Keywords: aging; cell composition; epigenetic; epithelial cells; methylation; predictor; swab.
Conflict of interest statement
W.W. and U.G. are co-founders of Cygenia GmbH (
Figures
Similar articles
-
Aging of blood can be tracked by DNA methylation changes at just three CpG sites.Genome Biol. 2014 Feb 3;15(2):R24. doi: 10.1186/gb-2014-15-2-r24. Genome Biol. 2014. PMID: 24490752 Free PMC article.
-
Development of two age estimation models for buccal swab samples based on 3 CpG sites analyzed with pyrosequencing and minisequencing.Forensic Sci Int Genet. 2021 Jul;53:102521. doi: 10.1016/j.fsigen.2021.102521. Epub 2021 Apr 25. Forensic Sci Int Genet. 2021. PMID: 33933877
-
Postmortem age estimation via DNA methylation analysis in buccal swabs from corpses in different stages of decomposition-a "proof of principle" study.Int J Legal Med. 2021 Jan;135(1):167-173. doi: 10.1007/s00414-020-02360-7. Epub 2020 Jul 7. Int J Legal Med. 2021. PMID: 32632799 Free PMC article.
-
Forensic individual age estimation with DNA: From initial approaches to methylation tests.Forensic Sci Rev. 2017 Jul;29(2):121-144. Forensic Sci Rev. 2017. PMID: 28691915 Review.
-
Do age-associated DNA methylation changes increase the risk of malignant transformation?Bioessays. 2015 Jan;37(1):20-4. doi: 10.1002/bies.201400063. Epub 2014 Oct 9. Bioessays. 2015. PMID: 25303747 Review.
Cited by
-
New targeted approaches for epigenetic age predictions.BMC Biol. 2020 Jun 24;18(1):71. doi: 10.1186/s12915-020-00807-2. BMC Biol. 2020. PMID: 32580727 Free PMC article.
-
Early-life adversity and neurological disease: age-old questions and novel answers.Nat Rev Neurol. 2019 Nov;15(11):657-669. doi: 10.1038/s41582-019-0246-5. Epub 2019 Sep 17. Nat Rev Neurol. 2019. PMID: 31530940 Free PMC article. Review.
-
Rapamycin retards epigenetic ageing of keratinocytes independently of its effects on replicative senescence, proliferation and differentiation.Aging (Albany NY). 2019 May 26;11(10):3238-3249. doi: 10.18632/aging.101976. Aging (Albany NY). 2019. PMID: 31136303 Free PMC article.
-
Childhood environment influences epigenetic age and methylation concordance of a CpG clock locus in British-Bangladeshi migrants.Epigenetics. 2023 Dec;18(1):2153511. doi: 10.1080/15592294.2022.2153511. Epub 2022 Dec 10. Epigenetics. 2023. PMID: 36495138 Free PMC article.
-
Integration of DNA methylation patterns and genetic variation in human pediatric tissues help inform EWAS design and interpretation.Epigenetics Chromatin. 2019 Jan 2;12(1):1. doi: 10.1186/s13072-018-0245-6. Epigenetics Chromatin. 2019. PMID: 30602389 Free PMC article.
References
-
- Ritz-Timme S, Cattaneo C, Collins MJ, Waite ER, Schutz HW, Kaatsch HJ, Borrman HI. Age estimation: the state of the art in relation to the specific demands of forensic practise. Int J Legal Med. 2000;113:129–36. - PubMed
-
- Tsuji A, Ishiko A, Takasaki T, Ikeda N. Estimating age of humans based on telomere shortening. Forensic Sci Int. 2002;126:197–99. - PubMed
-
- Zubakov D, Liu F, van Zelm MC, Vermeulen J, Oostra BA, van Duijn CM, Driessen GJ, van Dongen JJ, Kayser M, Langerak AW. Estimating human age from T-cell DNA rearrangements. Curr Biol. 2010;20:970–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

