Recent advances in dynamic m6A RNA modification
- PMID: 27249342
- PMCID: PMC4852458
- DOI: 10.1098/rsob.160003
Recent advances in dynamic m6A RNA modification
Abstract
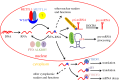
The identification of m(6)A demethylases and high-throughput sequencing analysis of methylated transcriptome corroborated m(6)A RNA epigenetic modification as a dynamic regulation process, and reignited its investigation in the past few years. Many basic concepts of cytogenetics have been revolutionized by the growing understanding of the fundamental role of m(6)A in RNA splicing, degradation and translation. In this review, we summarize typical features of methylated transcriptome in mammals, and highlight the 'writers', 'erasers' and 'readers' of m(6)A RNA modification. Moreover, we emphasize recent advances of biological functions of m(6)A and conceive the possible roles of m(6)A in the regulation of immune response and related diseases.
Keywords: m6A; mRNA; methylation.
© 2016 The Authors.
Figures
References
-
- Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. 2013. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 342, 1100–1104. (doi:10.1126/science.1242379) - DOI - PMC - PubMed
-
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. 2013. Variation and genetic control of protein abundance in humans. Nature 499, 79–82. (doi:10.1038/nature12223) - DOI - PMC - PubMed
-
- Motorin Y, Helm M. 2011. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631. (doi:10.1002/wrna.79) - DOI - PubMed
-
- Machnicka MA, et al. 2013. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 41, D262–D267. (doi:10.1093/nar/gks1007) - DOI - PMC - PubMed
-
- Desrosiers R, Friderici K, Rottman F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. USA 71, 3971–3975. (doi:10.1073/pnas.71.10.3971) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
