Non-Metabolic Role of PKM2 in Regulation of the HIV-1 LTR
- PMID: 27249540
- PMCID: PMC5714288
- DOI: 10.1002/jcp.25445
Non-Metabolic Role of PKM2 in Regulation of the HIV-1 LTR
Abstract
Identification of cellular proteins, in addition to already known transcription factors such as NF-κB, Sp1, C-EBPβ, NFAT, ATF/CREB, and LEF-1, which interact with the HIV-1 LTR, is critical in understanding the mechanism of HIV-1 replication in monocytes/macrophages. Our studies demonstrate upregulation of pyruvate kinase isoform M2 (PKM2) expression during HIV-1SF162 infection of monocyte/macrophages and reactivation of HIV-1 in U1 cells, a macrophage model of latency. We observed that HIV-1SF162 infection of monocyte/macrophages and reactivation of HIV-1 in U1 cells by PMA resulted in increased levels of nuclear PKM2 compared to PMA-induced U937 cells. Furthermore, there was a significant increase in the nuclear dimeric form of PKM2 in the PMA-induced U1 cells in comparison to PMA-induced U937 cells. We focused on understanding the potential role of PKM2 in HIV-1 LTR transactivation. Chromatin immunoprecipitation (ChIP) analysis in PMA-activated U1 and TZM-bl cells demonstrated the interaction of PKM2 with the HIV-1 LTR. Our studies show that overexpression of PKM2 results in transactivation of HIV-1 LTR-luciferase reporter in U937, U-87 MG, and TZM-bl cells. Using various truncated constructs of the HIV-1 LTR, we mapped the region spanning -120 bp to -80 bp to be essential for PKM2-mediated transactivation. This region contains the NF-κB binding site and deletion of this site attenuated PKM2-mediated activation of HIV-1 LTR. Immunoprecipitation experiments using U1 cell lysates demonstrated a physical interaction between PKM2 and the p65 subunit of NF-κB. These observations demonstrate for the first time that PKM2 is a transcriptional co-activator of HIV-1 LTR. J. Cell. Physiol. 232: 517-525, 2017. © 2016 Wiley Periodicals, Inc.
© 2016 Wiley Periodicals, Inc.
Figures


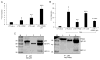

Similar articles
-
Negative regulation of HIV-1 transcription by a heterodimeric NF-κB1/p50 and C-terminally truncated STAT5 complex.J Mol Biol. 2011 Jul 29;410(5):933-43. doi: 10.1016/j.jmb.2011.03.044. J Mol Biol. 2011. PMID: 21763497
-
Poly (ADP-ribose) polymerase is involved in PMA-induced activation of HIV-1 in U1 cells by modulating the LTR function.Biochem Biophys Res Commun. 1999 Aug 19;262(1):285-9. doi: 10.1006/bbrc.1999.1146. Biochem Biophys Res Commun. 1999. PMID: 10448106
-
Analysis of the HIV-1 LTR NF-kappaB-proximal Sp site III: evidence for cell type-specific gene regulation and viral replication.Virology. 2000 Sep 1;274(2):262-77. doi: 10.1006/viro.2000.0476. Virology. 2000. PMID: 10964770
-
[Molecular pathogenesis in tuberculosis complicated with AIDS].Kekkaku. 2004 Nov;79(11):659-67. Kekkaku. 2004. PMID: 15729891 Review. Japanese.
-
HIV UTR, LTR, and Epigenetic Immunity.Viruses. 2022 May 18;14(5):1084. doi: 10.3390/v14051084. Viruses. 2022. PMID: 35632825 Free PMC article. Review.
Cited by
-
PKM2 deficiency exacerbates gram-negative sepsis-induced cardiomyopathy via disrupting cardiac calcium homeostasis.Cell Death Discov. 2022 Dec 23;8(1):496. doi: 10.1038/s41420-022-01287-9. Cell Death Discov. 2022. PMID: 36564378 Free PMC article.
-
PKM2 Is Required to Activate Myeloid Dendritic Cells from Patients with Severe Aplastic Anemia.Oxid Med Cell Longev. 2018 Feb 15;2018:1364165. doi: 10.1155/2018/1364165. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 29636835 Free PMC article.
References
-
- Ahmad N, Venkatesan S. Nef protein of HIV-1 is a transcriptional repressor of HIV-1 LTR. Science. 1988;241:1481–1485. - PubMed
-
- Amini S, Clavo A, Nadraga Y, Giordano A, Khalili K, Sawaya BE. Interplay between cdk9 and NF-kappaB factors determines the level of HIV-1 gene transcription in astrocytic cells. Oncogene. 2002;21:5797–5803. - PubMed
-
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. - PMC - PubMed
-
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

