High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii
- PMID: 27250364
- PMCID: PMC4890121
- DOI: 10.1038/srep27289
High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii
Abstract
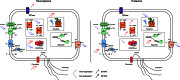
Pyrococcus yayanosii CH1, as the first and only obligate piezophilic hyperthermophilic microorganism discovered to date, extends the physical and chemical limits of life on Earth. It was isolated from the Ashadze hydrothermal vent at 4,100 m depth. Multi-omics analyses were performed to study the mechanisms used by the cell to cope with high hydrostatic pressure variations. In silico analyses showed that the P. yayanosii genome is highly adapted to its harsh environment, with a loss of aromatic amino acid biosynthesis pathways and the high constitutive expression of the energy metabolism compared with other non-obligate piezophilic Pyrococcus species. Differential proteomics and transcriptomics analyses identified key hydrostatic pressure-responsive genes involved in translation, chemotaxis, energy metabolism (hydrogenases and formate metabolism) and Clustered Regularly Interspaced Short Palindromic Repeats sequences associated with Cellular apoptosis susceptibility proteins.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Oger P. M. & Jebbar M. The many ways of coping with pressure. Res Microbiol 161, 799–809 (2010). - PubMed
-
- Abe F. & Horikoshi K. The biotechnological potential of piezophiles. Trends Biotechnol 19, 102–108 (2001). - PubMed
-
- Cario A., Lormières F., Xiang X. & Oger P. High hydrostatic pressure increases amino acid requirements in the piezo-hyperthermophilic archaeon Thermococcus barophilus. Res Microbiol 166, 710–716 (2015). - PubMed
-
- Vannier P., Michoud G., Oger P., Thór Marteinsson V. & Jebbar M. Genome expression of Thermococcus barophilus and Thermococcus kodakarensis in response to different hydrostatic pressure conditions. Res Microbiol 166, 717–725 (2015). - PubMed
-
- Simonato F. et al. Piezophilic adaptation: a genomic point of view. J Biotechnol 126, 11–25 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

