Proteogenomics connects somatic mutations to signalling in breast cancer
- PMID: 27251275
- PMCID: PMC5102256
- DOI: 10.1038/nature18003
Proteogenomics connects somatic mutations to signalling in breast cancer
Abstract
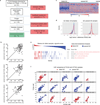
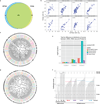
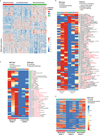
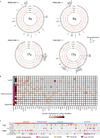
Somatic mutations have been extensively characterized in breast cancer, but the effects of these genetic alterations on the proteomic landscape remain poorly understood. Here we describe quantitative mass-spectrometry-based proteomic and phosphoproteomic analyses of 105 genomically annotated breast cancers, of which 77 provided high-quality data. Integrated analyses provided insights into the somatic cancer genome including the consequences of chromosomal loss, such as the 5q deletion characteristic of basal-like breast cancer. Interrogation of the 5q trans-effects against the Library of Integrated Network-based Cellular Signatures, connected loss of CETN3 and SKP1 to elevated expression of epidermal growth factor receptor (EGFR), and SKP1 loss also to increased SRC tyrosine kinase. Global proteomic data confirmed a stromal-enriched group of proteins in addition to basal and luminal clusters, and pathway analysis of the phosphoproteome identified a G-protein-coupled receptor cluster that was not readily identified at the mRNA level. In addition to ERBB2, other amplicon-associated highly phosphorylated kinases were identified, including CDK12, PAK1, PTK2, RIPK2 and TLK2. We demonstrate that proteogenomic analysis of breast cancer elucidates the functional consequences of somatic mutations, narrows candidate nominations for driver genes within large deletions and amplified regions, and identifies therapeutic targets.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
- U24 CA160034/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- U24 CA159988/CA/NCI NIH HHS/United States
- R00 CA166228/CA/NCI NIH HHS/United States
- U24CA159988/CA/NCI NIH HHS/United States
- R01GM108711/GM/NIGMS NIH HHS/United States
- RR140033/RR/NCRR NIH HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- P30 CA91842/CA/NCI NIH HHS/United States
- P41 GM103422/GM/NIGMS NIH HHS/United States
- U24CA160036/CA/NCI NIH HHS/United States
- U24 CA160036/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U24 CA160019/CA/NCI NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- U24 CA160035/CA/NCI NIH HHS/United States
- U24CA160034/CA/NCI NIH HHS/United States
- U24CA160035/CA/NCI NIH HHS/United States
- P41 RR000954/RR/NCRR NIH HHS/United States
- T32 GM088129/GM/NIGMS NIH HHS/United States
- U24CA160019/CA/NCI NIH HHS/United States
- R01 GM108711/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

