The gene cortex controls mimicry and crypsis in butterflies and moths
- PMID: 27251285
- PMCID: PMC5094491
- DOI: 10.1038/nature17961
The gene cortex controls mimicry and crypsis in butterflies and moths
Abstract
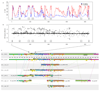
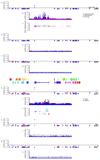
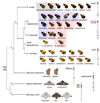
The wing patterns of butterflies and moths (Lepidoptera) are diverse and striking examples of evolutionary diversification by natural selection. Lepidopteran wing colour patterns are a key innovation, consisting of arrays of coloured scales. We still lack a general understanding of how these patterns are controlled and whether this control shows any commonality across the 160,000 moth and 17,000 butterfly species. Here, we use fine-scale mapping with population genomics and gene expression analyses to identify a gene, cortex, that regulates pattern switches in multiple species across the mimetic radiation in Heliconius butterflies. cortex belongs to a fast-evolving subfamily of the otherwise highly conserved fizzy family of cell-cycle regulators, suggesting that it probably regulates pigmentation patterning by regulating scale cell development. In parallel with findings in the peppered moth (Biston betularia), our results suggest that this mechanism is common within Lepidoptera and that cortex has become a major target for natural selection acting on colour and pattern variation in this group of insects.
Figures







References
-
- Jiggins CD. Ecological Speciation in Mimetic Butterflies. BioScience. 2008;58:541–548.
-
- Van’t Hof AE, et al. The industrial melanism mutation in British peppered moths is a transposable element. Nature. This issue. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/E006191/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H014268/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- H01439X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 243179/ERC_/European Research Council/International
- BB/H01439X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

