Discovery, characterization and in vivo activity of pyocin SD2, a protein antibiotic from Pseudomonas aeruginosa
- PMID: 27252387
- PMCID: PMC4964976
- DOI: 10.1042/BCJ20160470
Discovery, characterization and in vivo activity of pyocin SD2, a protein antibiotic from Pseudomonas aeruginosa
Abstract
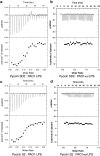
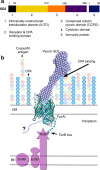
Increasing rates of antibiotic resistance among Gram-negative pathogens such as Pseudomonas aeruginosa means alternative approaches to antibiotic development are urgently required. Pyocins, produced by P. aeruginosa for intraspecies competition, are highly potent protein antibiotics known to actively translocate across the outer membrane of P. aeruginosa. Understanding and exploiting the mechanisms by which pyocins target, penetrate and kill P. aeruginosa is a promising approach to antibiotic development. In this work we show the therapeutic potential of a newly identified tRNase pyocin, pyocin SD2, by demonstrating its activity in vivo in a murine model of P. aeruginosa lung infection. In addition, we propose a mechanism of cell targeting and translocation for pyocin SD2 across the P. aeruginosa outer membrane. Pyocin SD2 is concentrated at the cell surface, via binding to the common polysaccharide antigen (CPA) of P. aeruginosa lipopolysaccharide (LPS), from where it can efficiently locate its outer membrane receptor FpvAI. This strategy of utilizing both the CPA and a protein receptor for cell targeting is common among pyocins as we show that pyocins S2, S5 and SD3 also bind to the CPA. Additional data indicate a key role for an unstructured N-terminal region of pyocin SD2 in the subsequent translocation of the pyocin into the cell. These results greatly improve our understanding of how pyocins target and translocate across the outer membrane of P. aeruginosa. This knowledge could be useful for the development of novel anti-pseudomonal therapeutics and will also support the development of pyocin SD2 as a therapeutic in its own right.
Keywords: Pseudomonas aeruginosa; antibiotics; bacteriocins; common polysaccharide antigen; outer membrane; pyocins.
© 2016 The Author(s).
Figures




References
-
- Cystic Fibrosis Trust. UK Cystic Fibrosis Registry Annual data report 2012. Cystic Fibrosis Trust; 2013.
-
- Sievert D.M., Ricks P., Edwards J.R., Schneider A., Patel J., Srinivasan A., Srinivasan A., Kallen A., Limbago B., Fridkin S. National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Cont. Hosp. Epidemiol. 2013;34:1–14. doi: 10.1086/668770. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

