Time-Dependent Rate Phenomenon in Viruses
- PMID: 27252529
- PMCID: PMC4984659
- DOI: 10.1128/JVI.00593-16
Time-Dependent Rate Phenomenon in Viruses
Abstract
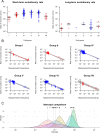
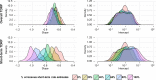
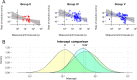
Among the most fundamental questions in viral evolutionary biology are how fast viruses evolve and how evolutionary rates differ among viruses and fluctuate through time. Traditionally, viruses are loosely classed into two groups: slow-evolving DNA viruses and fast-evolving RNA viruses. As viral evolutionary rate estimates become more available, it appears that the rates are negatively correlated with the measurement timescales and that the boundary between the rates of DNA and RNA viruses might not be as clear as previously thought. In this study, we collected 396 viral evolutionary rate estimates across almost all viral genome types and replication strategies, and we examined their rate dynamics. We showed that the time-dependent rate phenomenon exists across multiple levels of viral taxonomy, from the Baltimore classification viral groups to genera. We also showed that, by taking the rate decay dynamics into account, a clear division between the rates of DNA and RNA viruses as well as reverse-transcribing viruses could be recovered. Surprisingly, despite large differences in their biology, our analyses suggested that the rate decay speed is independent of viral types and thus might be useful for better estimation of the evolutionary time scale of any virus. To illustrate this, we used our model to reestimate the evolutionary timescales of extant lentiviruses, which were previously suggested to be very young by standard phylogenetic analyses. Our analyses suggested that these viruses are millions of years old, in agreement with paleovirological evidence, and therefore, for the first time, reconciled molecular analyses of ancient and extant viruses.
Importance: This work provides direct evidence that viral evolutionary rate estimates decay with their measurement timescales and that the rate decay speeds do not differ significantly among viruses despite the vast differences in their molecular features. After adjustment for the rate decay dynamics, the division between the rates of double-stranded DNA (dsDNA), single-stranded RNA (ssRNA), and ssDNA/reverse-transcribing viruses could be seen more clearly than before. Our results provide a guideline for further improvement of the molecular clock. As a demonstration of this, we used our model to reestimate the timescales of modern lentiviruses, which were previously thought to be very young, and concluded that they are millions of years old. This result matches the estimate from paleovirological analyses, thus bridging the gap between ancient and extant viral evolutionary studies.
Copyright © 2016 Aiewsakun and Katzourakis.
Figures


References
-
- Sakaoka H, Kurita K, Iida Y, Takada S, Umene K, Kim YT, Ren CS, Nahmias AJ. 1994. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J Gen Virol 75:513–527. doi: 10.1099/0022-1317-75-3-513. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

