Using the E4orf6-Based E3 Ubiquitin Ligase as a Tool To Analyze the Evolution of Adenoviruses
- PMID: 27252531
- PMCID: PMC4984651
- DOI: 10.1128/JVI.00420-16
Using the E4orf6-Based E3 Ubiquitin Ligase as a Tool To Analyze the Evolution of Adenoviruses
Abstract
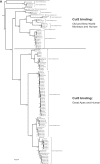
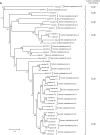
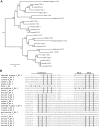
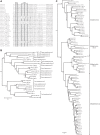
E4orf6 proteins from all human adenoviruses form Cullin-based ubiquitin ligase complexes that, in association with E1B55K, target cellular proteins for degradation. While most are assembled with Cul5, a few utilize Cul2. BC-box motifs enable all these E4orf6 proteins to assemble ligase complexes with Elongins B and C. We also identified a Cul2-box motif used for Cul2 selection in all Cul2-based complexes. With this information, we set out to determine if other adenoviruses also possess the ability to form the ligase complex and, if so, to predict their Cullin usage. Here we report that all adenoviruses known to encode an E4orf6-like protein (mastadenoviruses and atadenoviruses) maintain the potential to form the ligase complex. We could accurately predict Cullin usage for E4orf6 products of mastadenoviruses and all but one atadenovirus. Interestingly, in nonhuman primate adenoviruses, we found a clear segregation of Cullin binding, with Cul5 utilized by viruses infecting great apes and Cul2 by Old/New World monkey viruses, suggesting that a switch from Cul2 to Cul5 binding occurred during the period when great apes diverged from monkeys. Based on the analysis of Cullin selection, we also suggest that the majority of human adenoviruses, which exhibit a broader tropism for the eye and the respiratory tract, exhibit Cul5 specificity and resemble viruses infecting great apes, whereas those that infect the gastrointestinal tract may have originated from monkey viruses that share Cul2 specificity. Finally, aviadenoviruses also appear to contain E4orf6 genes that encode proteins with a conserved XCXC motif followed by, in most cases, a BC-box motif.
Importance: Two early adenoviral proteins, E4orf6 and E1B55K, form a ubiquitin ligase complex with cellular proteins to ubiquitinate specific substrates, leading to their degradation by the proteasome. In studies with representatives of each human adenovirus species, we (and others) previously discovered that some viruses use Cul2 to form the complex, while others use Cul5. In the present study, we expanded our analyses to all sequenced adenoviruses and found that E4orf6 genes from all mast- and atadenoviruses encode proteins containing the motifs necessary to form the ligase complex. We found a clear separation in Cullin specificity between adenoviruses of great apes and Old/New World monkeys, lending support for a monkey origin for human viruses of the Human mastadenovirus A, F, and G species. We also identified previously unrecognized E4orf6 genes in the aviadenoviruses that encode proteins containing motifs permitting formation of the ubiquitin ligase.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Benko M, Harrach B. 2003. Molecular evolution of adenoviruses. Curr Top Microbiol Immunol 272:3–35. - PubMed
-
- Shenk T. 1989. Oncogenesis by DNA viruses: adenovirus, p 239–257. In Weinberg RA. (ed), Oncogenes and the molecular origins of cancer, vol 18 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

