Community Structure Reveals Biologically Functional Modules in MEF2C Transcriptional Regulatory Network
- PMID: 27252657
- PMCID: PMC4878384
- DOI: 10.3389/fphys.2016.00184
Community Structure Reveals Biologically Functional Modules in MEF2C Transcriptional Regulatory Network
Abstract
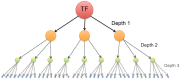
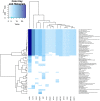
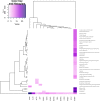
Gene regulatory networks are useful to understand the activity behind the complex mechanisms in transcriptional regulation. A main goal in contemporary biology is using such networks to understand the systemic regulation of gene expression. In this work, we carried out a systematic study of a transcriptional regulatory network derived from a comprehensive selection of all potential transcription factor interactions downstream from MEF2C, a human transcription factor master regulator. By analyzing the connectivity structure of such network, we were able to find different biologically functional processes and specific biochemical pathways statistically enriched in communities of genes into the network, such processes are related to cell signaling, cell cycle and metabolism. In this way we further support the hypothesis that structural properties of biological networks encode an important part of their functional behavior in eukaryotic cells.
Keywords: FANTOM4; InfoMap; MEF2C; community structure; transcription factor; transcriptional regulatory networks.
Figures







References
-
- Barabási A.-L., Albert R., Jeong H. (2000). Scale-free characteristics of random networks: the topology of the world-wide web. Physica A 281, 69–77. 10.1016/S0378-4371(00)00018-2 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

