Metagenomic Reconstruction of Key Anaerobic Digestion Pathways in Municipal Sludge and Industrial Wastewater Biogas-Producing Systems
- PMID: 27252693
- PMCID: PMC4879347
- DOI: 10.3389/fmicb.2016.00778
Metagenomic Reconstruction of Key Anaerobic Digestion Pathways in Municipal Sludge and Industrial Wastewater Biogas-Producing Systems
Abstract
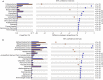
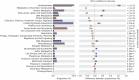
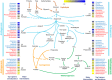
Anaerobic digestion (AD) is a microbial process widely used to treat organic wastes. While the microbes involved in digestion of municipal sludge are increasingly well characterized, the taxonomic and functional compositions of AD digesters treating industrial wastewater have been understudied. This study examined metagenomes from a biogas-producing digester treating municipal sludge in Shek Wu Hui (SWH), Hong Kong and an industrial wastewater digester in Guangzhou (GZ), China, and compared their taxonomic composition and reconstructed biochemical pathways. Genes encoding carbohydrate metabolism and protein metabolism functions were overrepresented in GZ, while genes encoding functions related to fatty acids, lipids and isoprenoids were overrepresented in SWH, reflecting the plants' feedstocks. Mapping of genera to functions in each community indicated that both digesters had a high level of functional redundancy, and a more even distribution of genera in GZ suggested that it was more functionally stable. While fermentation in both samples was dominated by Clostridia, SWH had an overrepresentation of Proteobacteria, including syntrophic acetogens, reflecting its more complex substrate. Considering the growing importance of biogas as an alternative fuel source, a detailed mechanistic understanding of AD is important and this report will be a basis for further study of industrial wastewater AD.
Keywords: anaerobic digestion; biogas; industrial wastewater; metagenomes; municipal sludge.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

