A 3D in vitro model to explore the inter-conversion between epithelial and mesenchymal states during EMT and its reversion
- PMID: 27255191
- PMCID: PMC4891772
- DOI: 10.1038/srep27072
A 3D in vitro model to explore the inter-conversion between epithelial and mesenchymal states during EMT and its reversion
Abstract
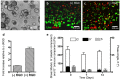
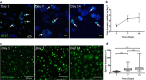
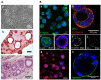
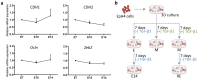
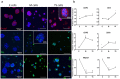
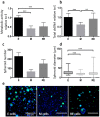
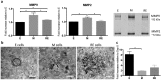
Epithelial-to-mesenchymal transitions (EMT) are strongly implicated in cancer dissemination. Intermediate states, arising from inter-conversion between epithelial (E) and mesenchymal (M) states, are characterized by phenotypic heterogeneity combining E and M features and increased plasticity. Hybrid EMT states are highly relevant in metastatic contexts, but have been largely neglected, partially due to the lack of physiologically-relevant 3D platforms to study them. Here we propose a new in vitro model, combining mammary E cells with a bioengineered 3D matrix, to explore phenotypic and functional properties of cells in transition between E and M states. Optimized alginate-based 3D matrices provided adequate 3D microenvironments, where normal epithelial morphogenesis was recapitulated, with formation of acini-like structures, similar to those found in native mammary tissue. TGFβ1-driven EMT in 3D could be successfully promoted, generating M-like cells. TGFβ1 removal resulted in phenotypic switching to an intermediate state (RE cells), a hybrid cell population expressing both E and M markers at gene/protein levels. RE cells exhibited increased proliferative/clonogenic activity, as compared to M cells, being able to form large colonies containing cells with front-back polarity, suggesting a more aggressive phenotype. Our 3D model provides a powerful tool to investigate the role of the microenvironment on metastable EMT stages.
Figures
References
-
- Yang J. & Weinberg R. A. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Dev Cell 14, 818–829 (2008). - PubMed
-
- Thiery J. P. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2, 442–454 (2002). - PubMed
-
- Brabletz T. EMT and MET in Metastasis: Where Are the Cancer Stem Cells? Cancer Cell 22, 699–701 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

