Synthetic mixed-signal computation in living cells
- PMID: 27255669
- PMCID: PMC4895730
- DOI: 10.1038/ncomms11658
Synthetic mixed-signal computation in living cells
Abstract
Living cells implement complex computations on the continuous environmental signals that they encounter. These computations involve both analogue- and digital-like processing of signals to give rise to complex developmental programs, context-dependent behaviours and homeostatic activities. In contrast to natural biological systems, synthetic biological systems have largely focused on either digital or analogue computation separately. Here we integrate analogue and digital computation to implement complex hybrid synthetic genetic programs in living cells. We present a framework for building comparator gene circuits to digitize analogue inputs based on different thresholds. We then demonstrate that comparators can be predictably composed together to build band-pass filters, ternary logic systems and multi-level analogue-to-digital converters. In addition, we interface these analogue-to-digital circuits with other digital gene circuits to enable concentration-dependent logic. We expect that this hybrid computational paradigm will enable new industrial, diagnostic and therapeutic applications with engineered cells.
Conflict of interest statement
J.R.R., G.S. and T.K.L. have filed a provisional patent application based on this work (‘Analogue to Digital Computations in Biological Systems', PCT/US2015/067381).
Figures
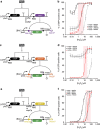
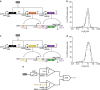

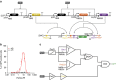
References
-
- Ausländer S., Ausländer D., Müller M., Wieland M. & Fussenegger M. Programmable single-cell mammalian biocomputers. Nature 487, 123–127 (2012). - PubMed
-
- Xie Z., Wroblewska L., Prochazka L., Weiss R. & Benenson Y. Multi-input RNAi-based logic circuit for indentification of specifc cancer cells. Science 333, 1307–1311 (2011). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

