MutSβ promotes trinucleotide repeat expansion by recruiting DNA polymerase β to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis
- PMID: 27255792
- PMCID: PMC5129881
- DOI: 10.1038/cr.2016.66
MutSβ promotes trinucleotide repeat expansion by recruiting DNA polymerase β to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis
Abstract
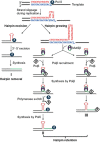
Expansion of (CAG)•(CTG) repeats causes a number of familial neurodegenerative disorders. Although the underlying mechanism remains largely unknown, components involved in DNA mismatch repair, particularly mismatch recognition protein MutSβ (a MSH2-MSH3 heterodimer), are implicated in (CAG)•(CTG) repeat expansion. In addition to recognizing small insertion-deletion loop-outs, MutSβ also specifically binds DNA hairpin imperfect heteroduplexes formed within (CAG)n•(CTG)n sequences. However, whether or not and how MutSβ binding triggers expansion of (CAG)•(CTG) repeats remain unknown. We show here that purified recombinant MutSβ physically interacts with DNA polymerase β (Polβ) and stimulates Polβ-catalyzed (CAG)n or (CTG)n hairpin retention. Consistent with these in vitro observations, MutSβ and Polβ interact with each other in vivo, and colocalize at (CAG)•(CTG) repeats during DNA replication. Our data support a model for error-prone processing of (CAG)n or (CTG)n hairpins by MutSβ and Polβ during DNA replication and/or repair: MutSβ recognizes (CAG)n or (CTG)n hairpins formed in the nascent DNA strand, and recruits Polβ to the complex, which then utilizes the hairpin as a primer for extension, leading to (CAG)•(CTG) repeat expansion. This study provides a novel mechanism for trinucleotide repeat expansion in both dividing and non-dividing cells.
Figures





Similar articles
-
Coordinated processing of 3' slipped (CAG)n/(CTG)n hairpins by DNA polymerases β and δ preferentially induces repeat expansions.J Biol Chem. 2013 May 24;288(21):15015-22. doi: 10.1074/jbc.M113.464370. Epub 2013 Apr 12. J Biol Chem. 2013. PMID: 23585564 Free PMC article.
-
Absence of MutSβ leads to the formation of slipped-DNA for CTG/CAG contractions at primate replication forks.DNA Repair (Amst). 2016 Jun;42:107-18. doi: 10.1016/j.dnarep.2016.04.002. Epub 2016 Apr 16. DNA Repair (Amst). 2016. PMID: 27155933 Free PMC article.
-
The Werner syndrome protein promotes CAG/CTG repeat stability by resolving large (CAG)(n)/(CTG)(n) hairpins.J Biol Chem. 2012 Aug 31;287(36):30151-6. doi: 10.1074/jbc.M112.389791. Epub 2012 Jul 11. J Biol Chem. 2012. PMID: 22787159 Free PMC article.
-
Disease-associated repeat instability and mismatch repair.DNA Repair (Amst). 2016 Feb;38:117-126. doi: 10.1016/j.dnarep.2015.11.008. Epub 2015 Dec 12. DNA Repair (Amst). 2016. PMID: 26774442 Review.
-
Coordinated roles of SLX4 and MutSβ in DNA repair and the maintenance of genome stability.Crit Rev Biochem Mol Biol. 2021 Apr;56(2):157-177. doi: 10.1080/10409238.2021.1881433. Epub 2021 Feb 17. Crit Rev Biochem Mol Biol. 2021. PMID: 33596761 Free PMC article. Review.
Cited by
-
Partners in crime: Tbf1 and Vid22 promote expansions of long human telomeric repeats at an interstitial chromosome position in yeast.PNAS Nexus. 2022 Jun 8;1(3):pgac080. doi: 10.1093/pnasnexus/pgac080. eCollection 2022 Jul. PNAS Nexus. 2022. PMID: 35832866 Free PMC article.
-
DNA mismatch repair in trinucleotide repeat instability.Sci China Life Sci. 2017 Oct;60(10):1087-1092. doi: 10.1007/s11427-017-9186-7. Epub 2017 Oct 24. Sci China Life Sci. 2017. PMID: 29075942 Free PMC article. Review.
-
Trinucleotide Repeat Expansion Diseases, RNAi, and Cancer.Trends Cancer. 2018 Oct;4(10):684-700. doi: 10.1016/j.trecan.2018.08.004. Epub 2018 Sep 26. Trends Cancer. 2018. PMID: 30292352 Free PMC article. Review.
-
Ethidium Bromide Modifies The Agarose Electrophoretic Mobility of CAG•CTG Alternative DNA Structures Generated by PCR.Front Cell Neurosci. 2017 May 30;11:153. doi: 10.3389/fncel.2017.00153. eCollection 2017. Front Cell Neurosci. 2017. PMID: 28611596 Free PMC article.
-
Replication stalling and DNA microsatellite instability.Biophys Chem. 2017 Jun;225:38-48. doi: 10.1016/j.bpc.2016.11.007. Epub 2016 Nov 22. Biophys Chem. 2017. PMID: 27914716 Free PMC article. Review.
References
-
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 2010; 11:165–170. - PubMed
-
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 2005; 6:729–742. - PubMed
-
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447:932–940. - PubMed
-
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 1995; 81:533–540. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

