C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector
- PMID: 27256883
- PMCID: PMC5127784
- DOI: 10.1126/science.aaf5573
C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector
Abstract
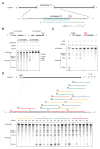
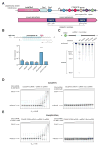
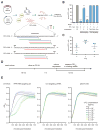
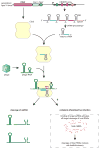
The clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated genes (Cas) adaptive immune system defends microbes against foreign genetic elements via DNA or RNA-DNA interference. We characterize the class 2 type VI CRISPR-Cas effector C2c2 and demonstrate its RNA-guided ribonuclease function. C2c2 from the bacterium Leptotrichia shahii provides interference against RNA phage. In vitro biochemical analysis shows that C2c2 is guided by a single CRISPR RNA and can be programmed to cleave single-stranded RNA targets carrying complementary protospacers. In bacteria, C2c2 can be programmed to knock down specific mRNAs. Cleavage is mediated by catalytic residues in the two conserved Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domains, mutations of which generate catalytically inactive RNA-binding proteins. These results broaden our understanding of CRISPR-Cas systems and suggest that C2c2 can be used to develop new RNA-targeting tools.
Copyright © 2016, American Association for the Advancement of Science.
Figures



References
-
- Wright AV, Nunez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164:29–44. - PubMed
-
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. - PubMed
-
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

