Mutation of the Traj18 gene segment using TALENs to generate Natural Killer T cell deficient mice
- PMID: 27256918
- PMCID: PMC4891675
- DOI: 10.1038/srep27375
Mutation of the Traj18 gene segment using TALENs to generate Natural Killer T cell deficient mice
Abstract
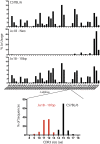
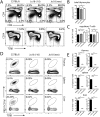
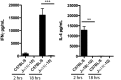
Invariant Natural Killer T (iNKT) cells are a unique subset of T lymphocytes that have been implicated in both promoting and suppressing a multitude of immune responses. In mice, iNKT cells express T cell antigen receptors (TCRs) comprising a unique TCRα rearrangement between the Trav11 and Traj18 gene segments. When paired with certain Trbv TCRβ chains, these TCRs recognize lipid antigens presented by the major histocompatibility complex (MHC) class I-like molecule, CD1d. Until recently, the sole model of iNKT deficiency targeted the Jα18, which is absolutely required to form the TCR with the appropriate antigenic specificity. However, these mice were demonstrated to have a large reduction in TCR repertoire diversity, which could confound results arising from studies using these mice. Here, we have created a new NKT-deficient mouse strain using transcription activator-like effector nuclease (TALEN) technology to only disrupt the expression of Jα18, leaving the remaining Jα repertoire unperturbed. We confirm that these mice lack iNKT cells and do not respond to lipid antigen stimulation while the development of conventional T cells, regulatory T cells, and type Ib NKT cells is normal. This new mouse strain will serve as a new model of iNKT cell deficiency to facilitate our understanding of iNKT biology.
Figures



Similar articles
-
New Genetically Manipulated Mice Provide Insights Into the Development and Physiological Functions of Invariant Natural Killer T Cells.Front Immunol. 2018 Jun 14;9:1294. doi: 10.3389/fimmu.2018.01294. eCollection 2018. Front Immunol. 2018. PMID: 29963043 Free PMC article. Review.
-
Mouse and Human CD1d-Self-Lipid Complexes Are Recognized Differently by Murine Invariant Natural Killer T Cell Receptors.PLoS One. 2016 May 23;11(5):e0156114. doi: 10.1371/journal.pone.0156114. eCollection 2016. PLoS One. 2016. PMID: 27213277 Free PMC article.
-
The ins and outs of type I iNKT cell development.Mol Immunol. 2019 Jan;105:116-130. doi: 10.1016/j.molimm.2018.09.023. Epub 2018 Nov 28. Mol Immunol. 2019. PMID: 30502719 Free PMC article. Review.
-
Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):E119-28. doi: 10.1073/pnas.1320777110. Epub 2013 Dec 16. Proc Natl Acad Sci U S A. 2014. PMID: 24344267 Free PMC article.
-
Immediate antigen-specific effector functions by TCR-transgenic CD8+ NKT cells.Eur J Immunol. 2006 Mar;36(3):570-82. doi: 10.1002/eji.200535461. Eur J Immunol. 2006. PMID: 16506291
Cited by
-
New Genetically Manipulated Mice Provide Insights Into the Development and Physiological Functions of Invariant Natural Killer T Cells.Front Immunol. 2018 Jun 14;9:1294. doi: 10.3389/fimmu.2018.01294. eCollection 2018. Front Immunol. 2018. PMID: 29963043 Free PMC article. Review.
-
Deficiency of Mucosal-Associated Invariant T Cells in TCRJα18 Germline Knockout Mice.Immunohorizons. 2019 Jun 11;3(6):203-207. doi: 10.4049/immunohorizons.1900035. Immunohorizons. 2019. PMID: 31356166 Free PMC article.
-
Differential Control of iNKT Cell Effector Lineage Differentiation by the Forkhead Box Protein O1 (Foxo1) Transcription Factor.Front Immunol. 2019 Nov 21;10:2710. doi: 10.3389/fimmu.2019.02710. eCollection 2019. Front Immunol. 2019. PMID: 31824499 Free PMC article.
-
Unveiling the heterogeneity of NKT cells in the liver through single cell RNA sequencing.Sci Rep. 2020 Nov 10;10(1):19453. doi: 10.1038/s41598-020-76659-1. Sci Rep. 2020. PMID: 33173202 Free PMC article.
-
Invariant Natural Killer T Cell Subsets-More Than Just Developmental Intermediates.Front Immunol. 2018 Jun 20;9:1393. doi: 10.3389/fimmu.2018.01393. eCollection 2018. Front Immunol. 2018. PMID: 29973936 Free PMC article. Review.
References
-
- Godfrey D. I., Uldrich A. P., McCluskey J., Rossjohn J. & Moody D. B. The burgeoning family of unconventional T cells. Nat Immunol 16, 1114–1123 (2015). - PubMed
-
- Godfrey D. I., MacDonald H. R., Kronenberg M., Smyth M. J. & Van Kaer L. NKT cells: what’s in a name? Nat Rev Immunol 4, 231–237 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

