RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals
- PMID: 27257063
- PMCID: PMC4994864
- DOI: 10.1093/nar/gkw508
RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals
Abstract
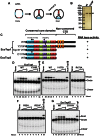
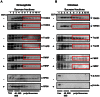
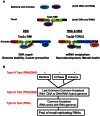
DNA Topoisomerases are essential to resolve topological problems during DNA metabolism in all species. However, the prevalence and function of RNA topoisomerases remain uncertain. Here, we show that RNA topoisomerase activity is prevalent in Type IA topoisomerases from bacteria, archaea, and eukarya. Moreover, this activity always requires the conserved Type IA core domains and the same catalytic residue used in DNA topoisomerase reaction; however, it does not absolutely require the non-conserved carboxyl-terminal domain (CTD), which is necessary for relaxation reactions of supercoiled DNA. The RNA topoisomerase activity of human Top3β differs from that of Escherichia coli topoisomerase I in that the former but not the latter requires the CTD, indicating that topoisomerases have developed distinct mechanisms during evolution to catalyze RNA topoisomerase reactions. Notably, Top3β proteins from several animals associate with polyribosomes, which are units of mRNA translation, whereas the Top3 homologs from E. coli and yeast lack the association. The Top3β-polyribosome association requires TDRD3, which directly interacts with Top3β and is present in animals but not bacteria or yeast. We propose that RNA topoisomerases arose in the early RNA world, and that they are retained through all domains of DNA-based life, where they mediate mRNA translation as part of polyribosomes in animals.
Published by Oxford University Press on behalf of Nucleic Acids Research 2016. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

