Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7
- PMID: 27257138
- PMCID: PMC4891710
- DOI: 10.1038/srep27060
Unexpected involvement of staple leads to redesign of selective bicyclic peptide inhibitor of Grb7
Abstract
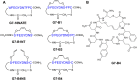
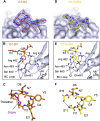
The design of potent and specific peptide inhibitors to therapeutic targets is of enormous utility for both proof-of-concept studies and for the development of potential new therapeutics. Grb7 is a key signaling molecule in the progression of HER2 positive and triple negative breast cancers. Here we report the crystal structure of a stapled bicyclic peptide inhibitor G7-B1 in complex with the Grb7-SH2 domain. This revealed an unexpected binding mode of the peptide, in which the staple forms an alternative contact with the surface of the target protein. Based on this structural information, we designed a new series of bicyclic G7 peptides that progressively constrain the starting peptide, to arrive at the G7-B4 peptide that binds with an approximately 2-fold enhanced affinity to the Grb7-SH2 domain (KD = 0.83 μM) compared to G7-B1 and shows low affinity binding to Grb2-, Grb10- and Grb14-SH2 domains (KD > 100 μM). Furthermore, we determined the structure of the G7-B4 bicyclic peptide in complex with the Grb7-SH2 domain, both before and after ring closing metathesis to show that the closed staple is essential to the target interaction. The G7-B4 peptide represents an advance in the development of Grb7 inhibitors and is a classical example of structure aided inhibitor development.
Figures



References
-
- Zinzalla G. & Thurston D. E. Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Future Med Chem 1, 65–93 (2009). - PubMed
-
- Benyamini H. & Friedler A. Using peptides to study protein-protein interactions. Future Med Chem 2, 989–1003 (2010). - PubMed
-
- Maes M., Loyter A. & Friedler A. Peptides that inhibit HIV-1 integrase by blocking its protein-protein interactions. FEBS J 279, 2795–2809 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

